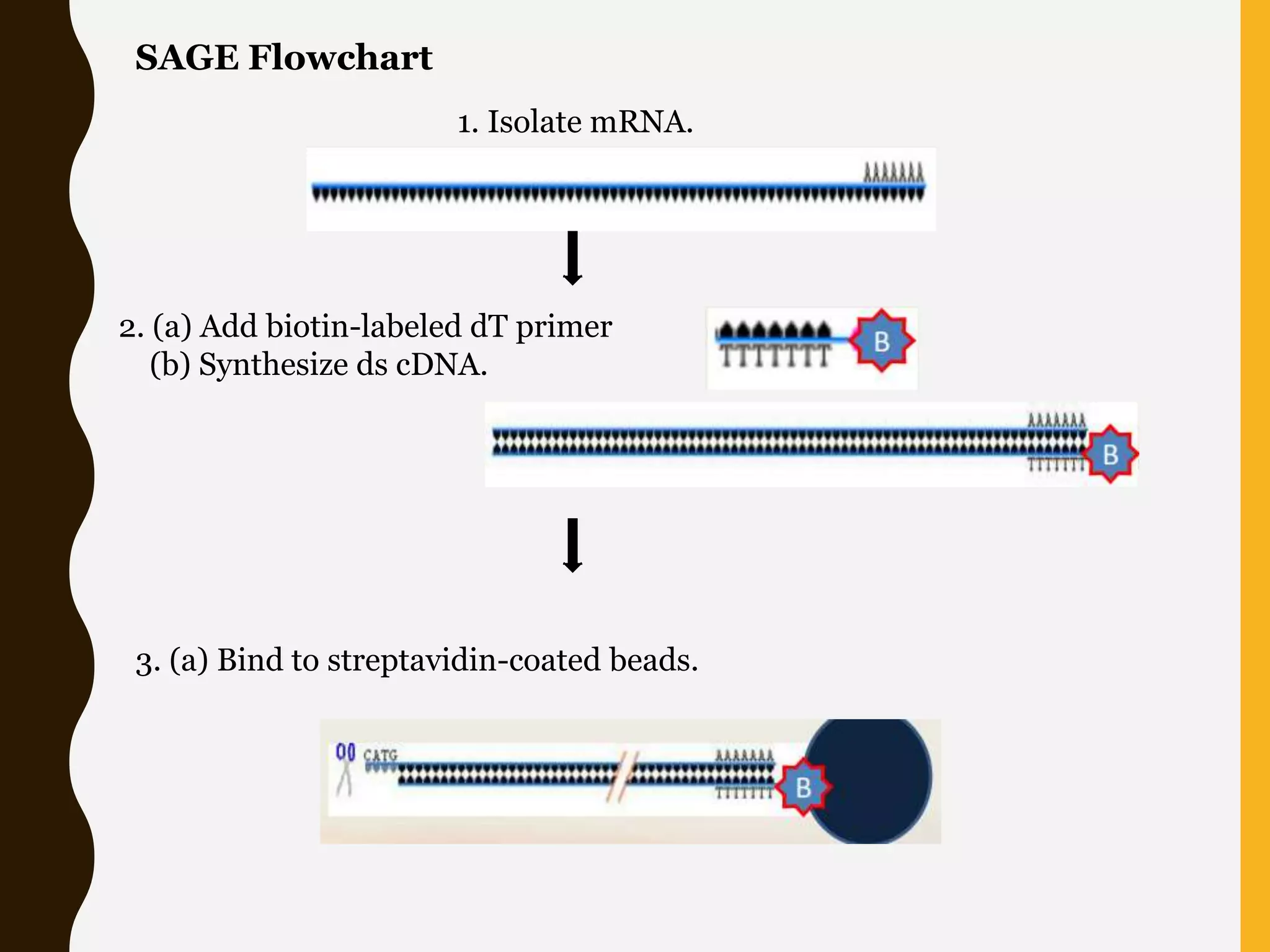
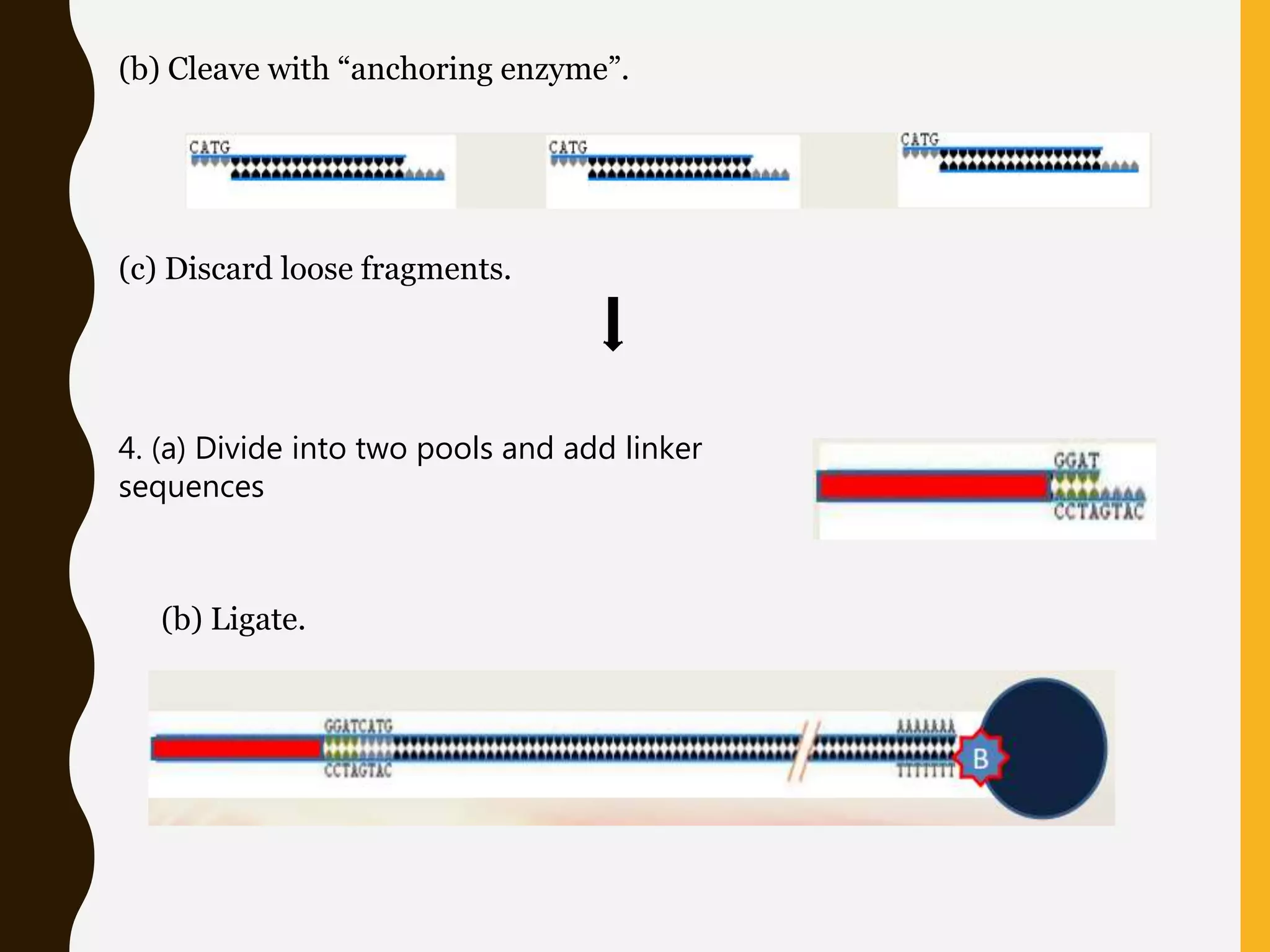
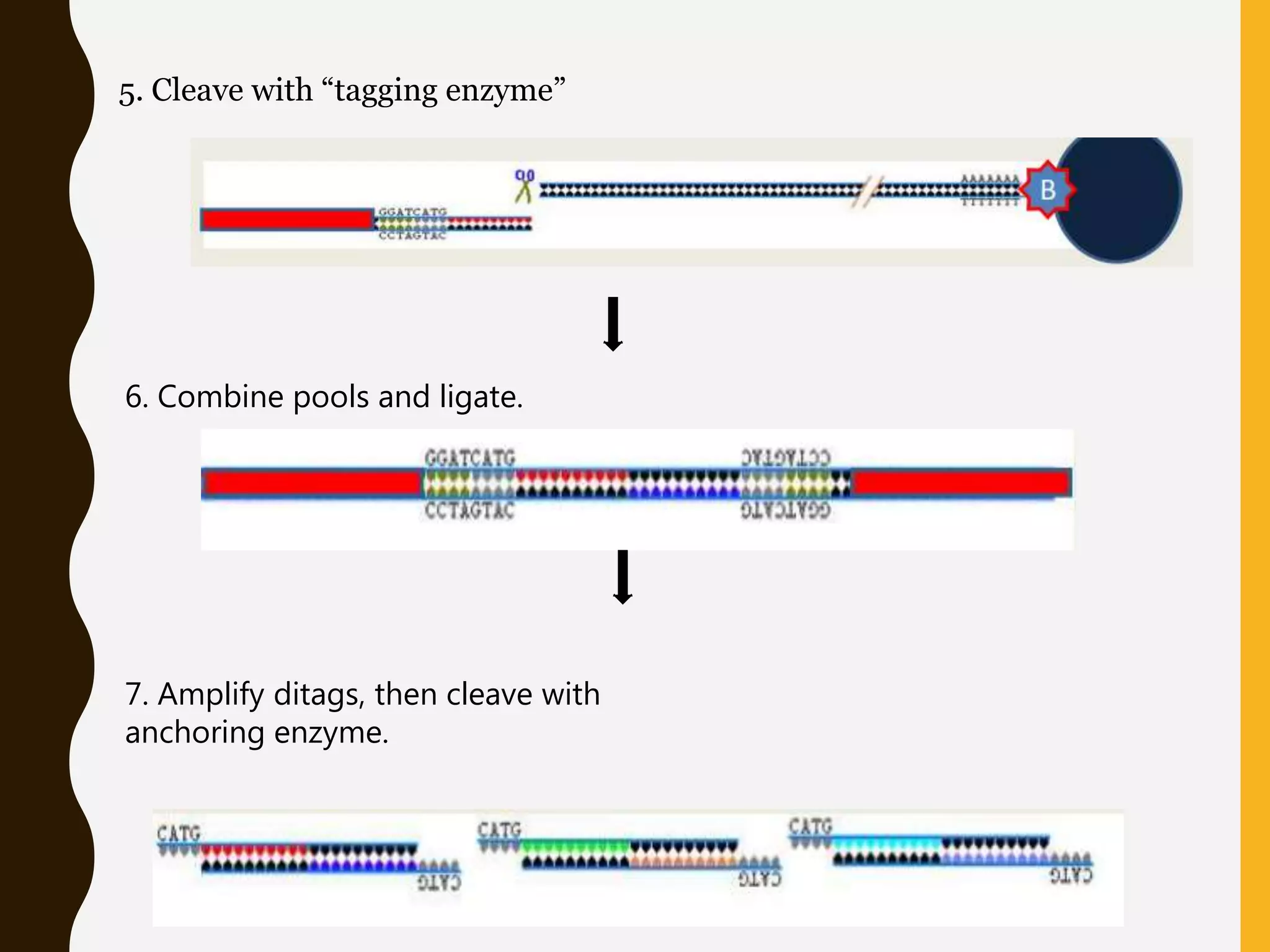
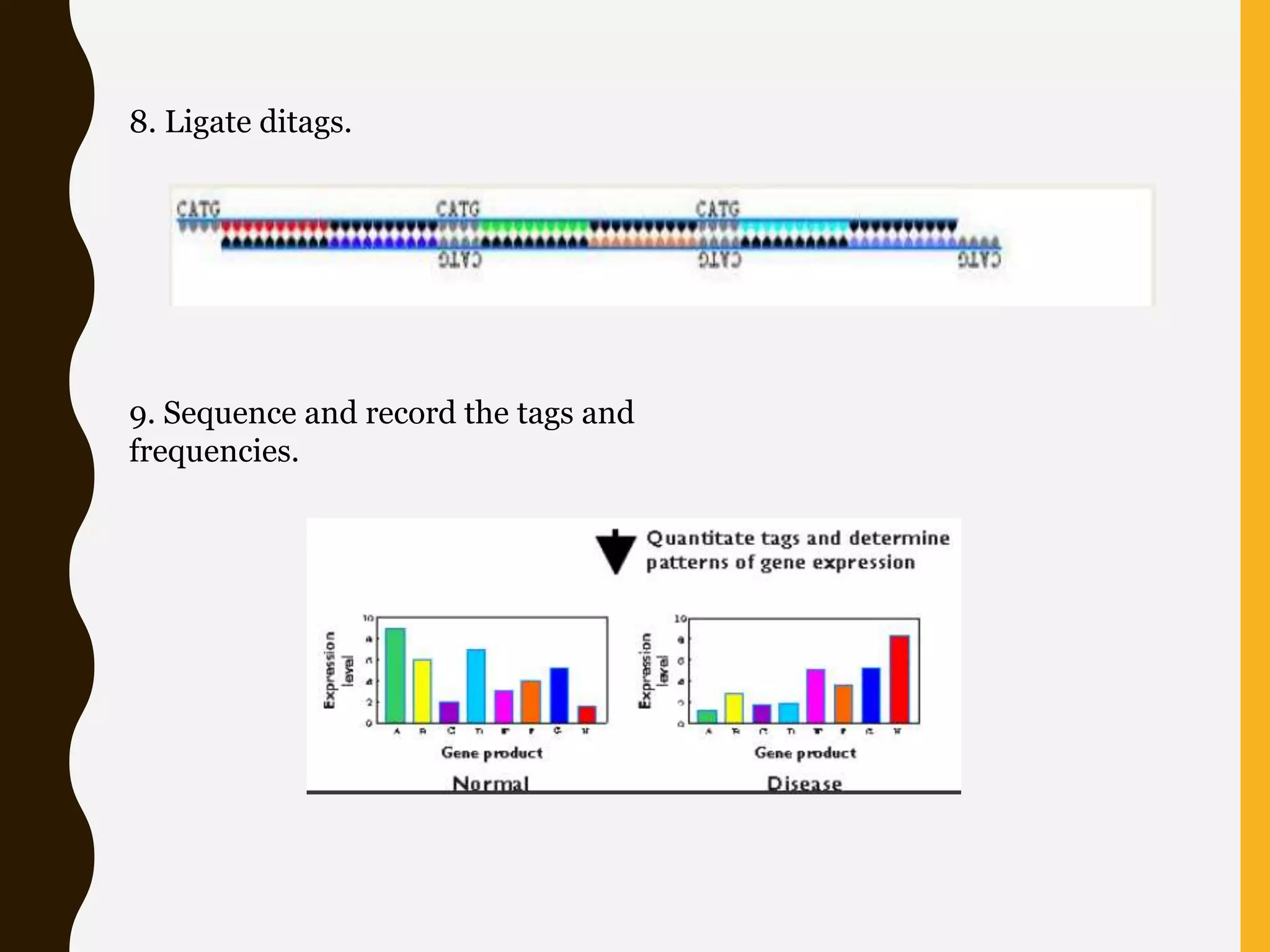
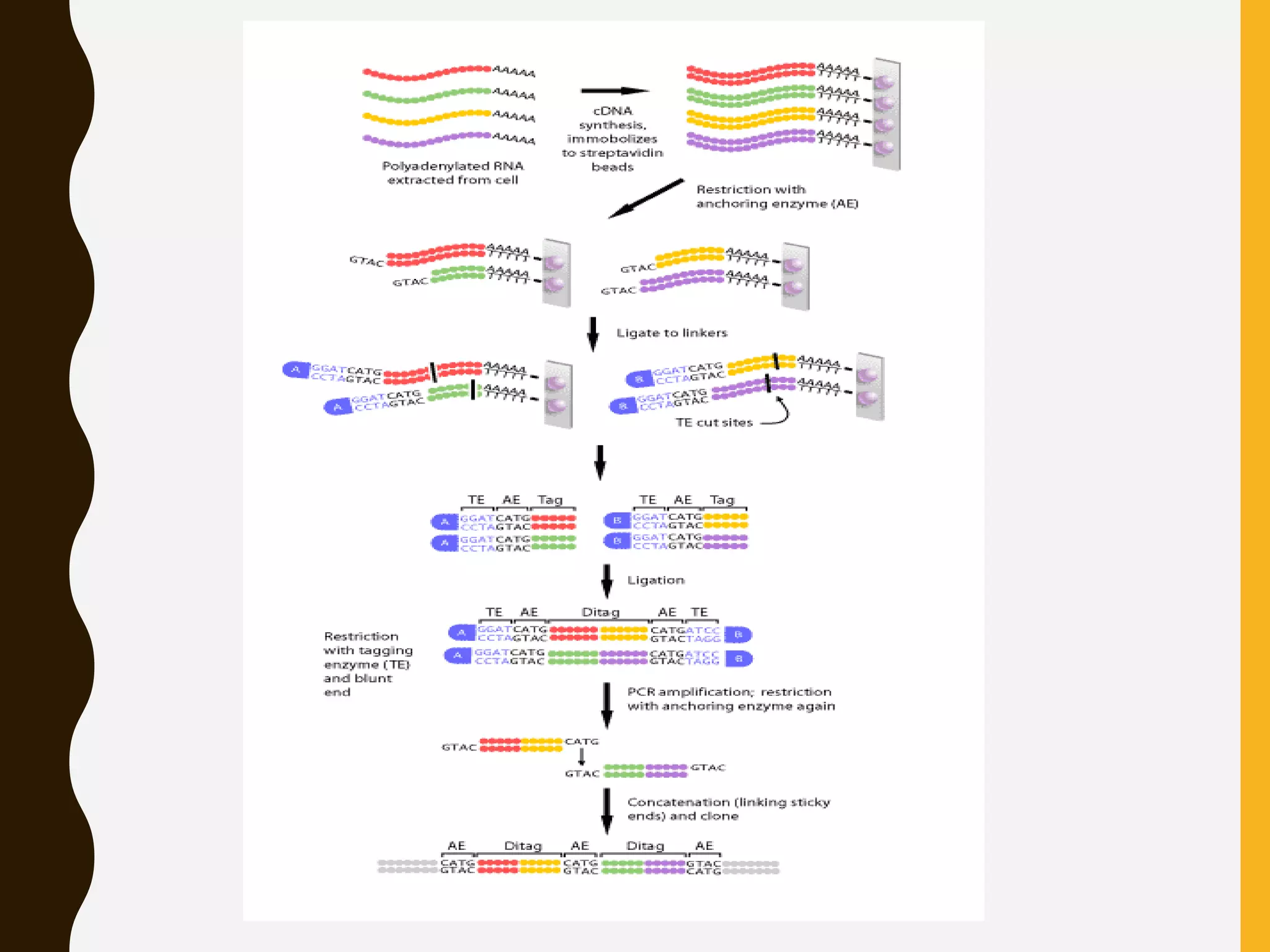
Serial Analysis of Gene Expression (SAGE) is a method that allows for the quantitative analysis of gene expression patterns in cells or tissues without prior knowledge of gene sequences. It works by extracting short sequence tags (10-14 base pairs) from the 3' end of transcripts and linking them together in long strings that can be cloned and sequenced simultaneously. This allows for many transcripts to be analyzed from a single sequencing reaction and provides quantitative data on gene expression levels based on tag frequencies. SAGE provides an accurate way to discover genes and analyze overall expression patterns without needing to know full mRNA sequences in advance.