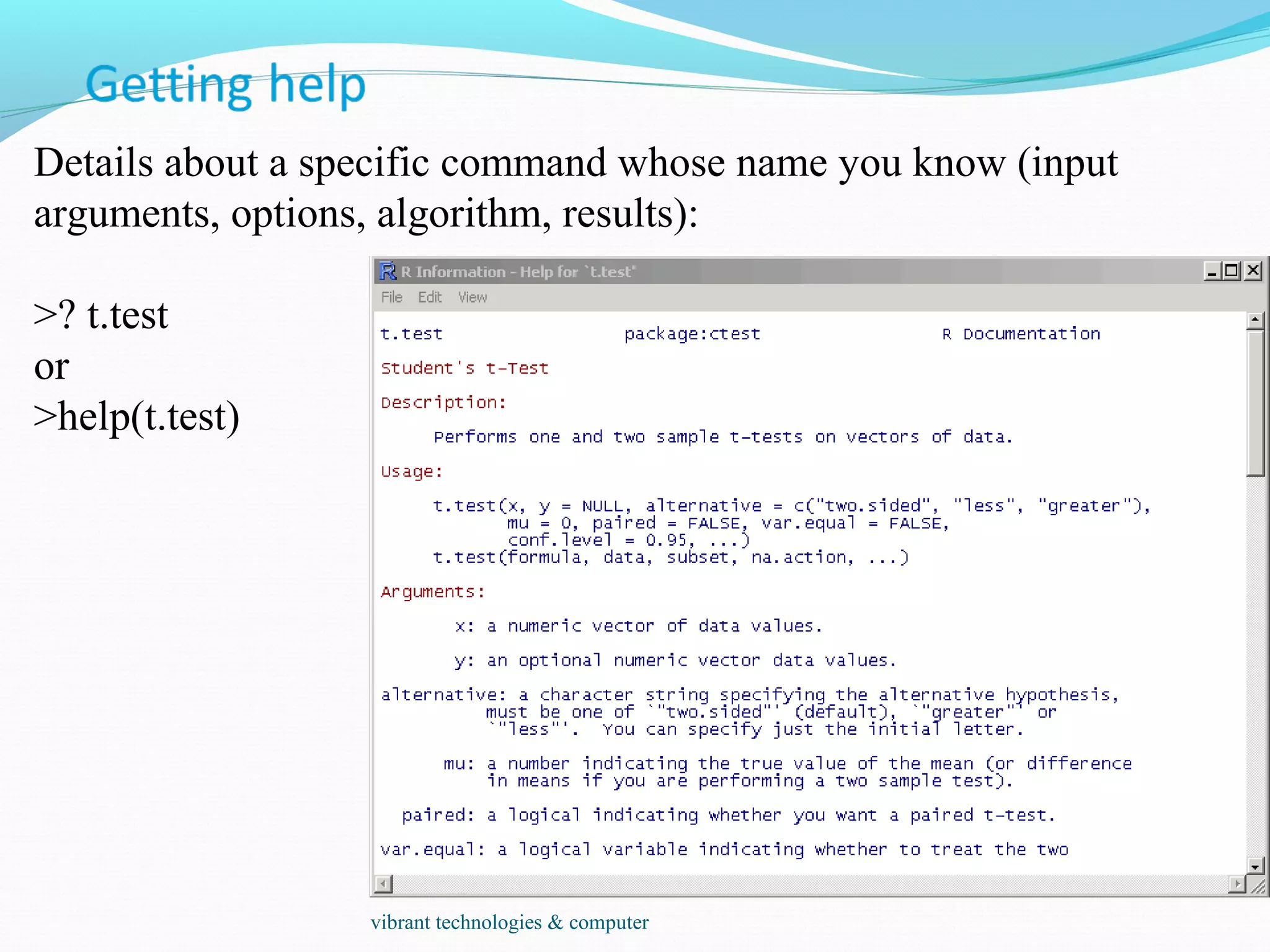
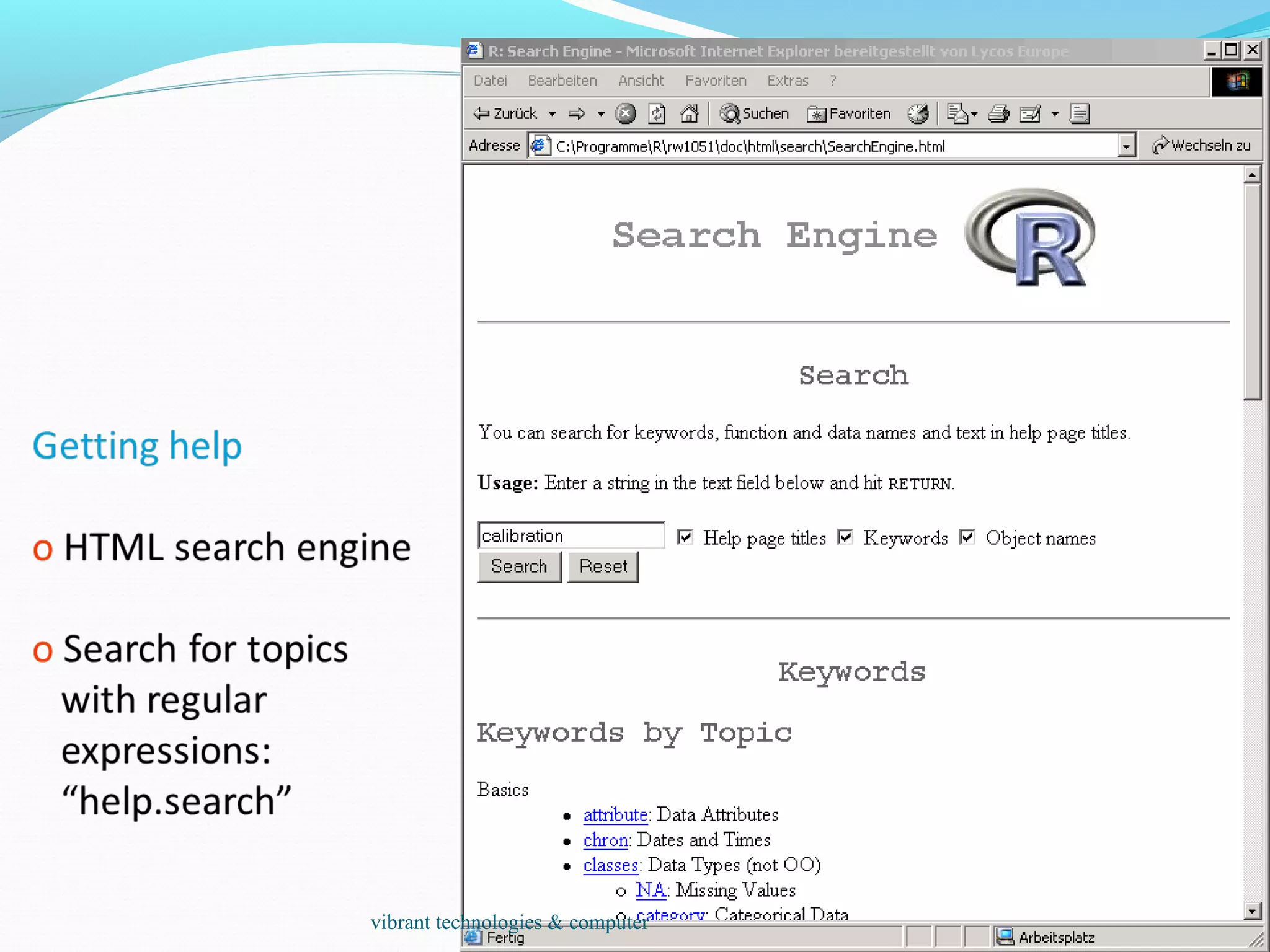
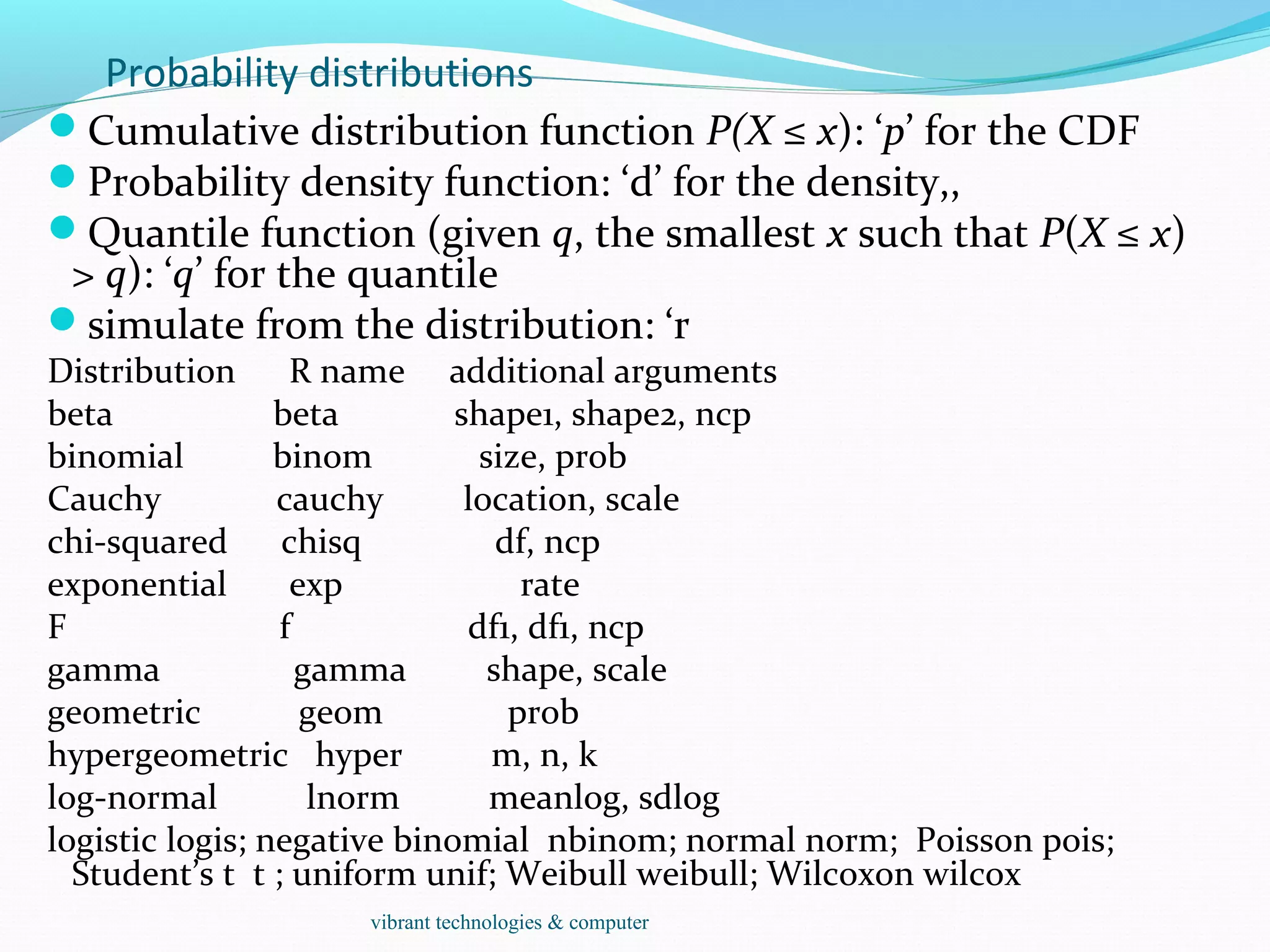
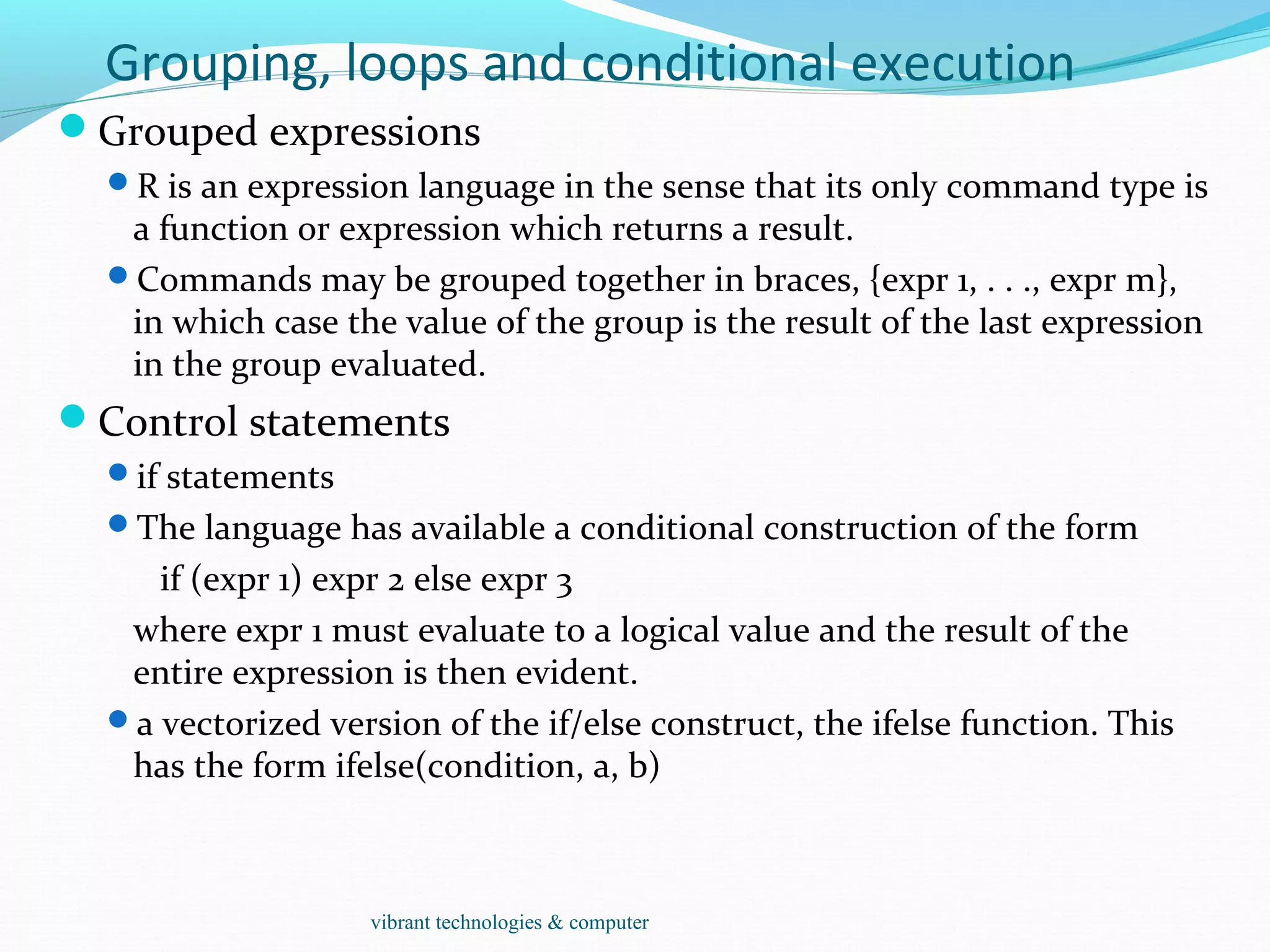
The document provides an outline of topics covered in R including introduction, data types, data analysis techniques like regression and ANOVA, resources for R, probability distributions, programming concepts like loops and functions, and data manipulation techniques. R is a programming language and software environment for statistical analysis that allows data manipulation, calculation, and graphical visualization. Key features of R include its programming language, high-level functions for statistics and graphics, and ability to extend functionality through packages.





![> log2(32)
[1] 5
> sqrt(2)
[1] 1.414214
> seq(0, 5, length=6)
[1] 0 1 2 3 4 5
> plot(sin(seq(0, 2*pi, length=100)))
0 20 40 60 80 100
-1.0-0.50.00.51.0
Index
sin(seq(0,2*pi,length=100))
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-9-2048.jpg)
![> a = 49
> sqrt(a)
[1] 7
> a = "The dog ate my homework"
> sub("dog","cat",a)
[1] "The cat ate my homework“
> a = (1+1==3)
> a
[1] FALSE
numeric
character
string
logical
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-13-2048.jpg)
![• vector: an ordered collection of data of the same type
> a = c(1,2,3)
> a*2
[1] 2 4 6
• Example: the mean spot intensities of all 15488 spots on a chip:
a vector of 15488 numbers
• In R, a single number is the special case of a vector with 1
element.
• Other vector types: character strings, logical
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-14-2048.jpg)
![• vector: an ordered collection of data of the same type.
> a = c(7,5,1)
> a[2]
[1] 5
• list: an ordered collection of data of arbitrary types.
> doe = list(name="john",age=28,married=F)
> doe$name
[1] "john“
> doe$age
[1] 28
• Typically, vector elements are accessed by their index (an integer),
list elements by their name (a character string). But both types
support both access methods.
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-16-2048.jpg)
![A character string can contain arbitrary text. Sometimes it is useful to use a limited
vocabulary, with a small number of allowed words. A factor is a variable that can only
take such a limited number of values, which are called levels.
> a
[1] Kolon(Rektum) Magen Magen
[4] Magen Magen Retroperitoneal
[7] Magen Magen(retrogastral) Magen
Levels: Kolon(Rektum) Magen Magen(retrogastral) Retroperitoneal
> class(a)
[1] "factor"
> as.character(a)
[1] "Kolon(Rektum)" "Magen" "Magen"
[4] "Magen" "Magen" "Retroperitoneal"
[7] "Magen" "Magen(retrogastral)" "Magen"
> as.integer(a)
[1] 1 2 2 2 2 4 2 3 2
> as.integer(as.character(a))
[1] NA NA NA NA NA NA NA NA NA NA NA NA
Warning message: NAs introduced by coercion
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-18-2048.jpg)
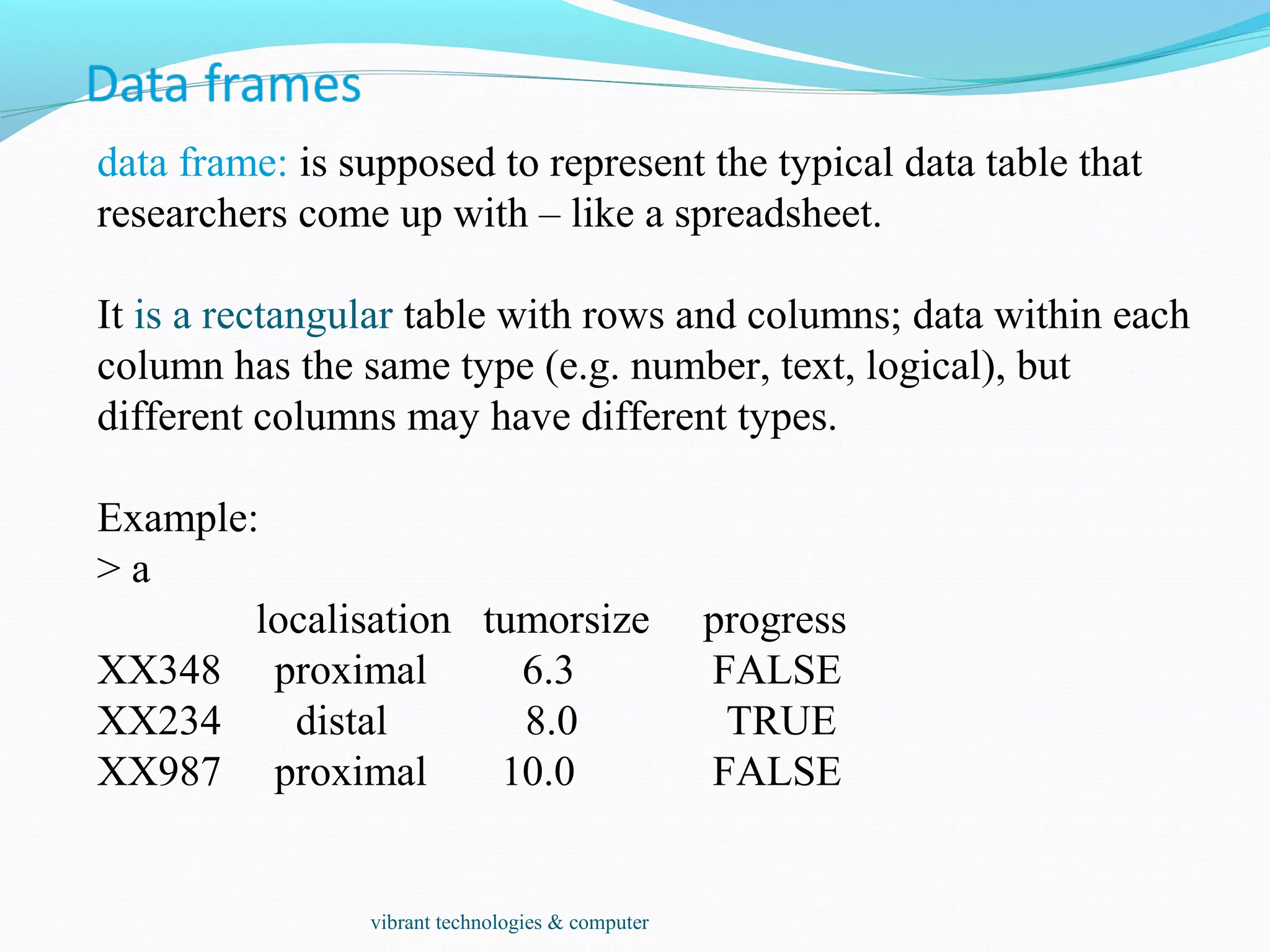
![Individual elements of a vector, matrix, array or data frame are
accessed with “[ ]” by specifying their index, or their name
> a
localisation tumorsize progress
XX348 proximal 6.3 0
XX234 distal 8.0 1
XX987 proximal 10.0 0
> a[3, 2]
[1] 10
> a["XX987", "tumorsize"]
[1] 10
> a["XX987",]
localisation tumorsize progress
XX987 proximal 10 0
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-19-2048.jpg)
![> a
localisation tumorsize progress
XX348 proximal 6.3 0
XX234 distal 8.0 1
XX987 proximal 10.0 0
> a[c(1,3),]
localisation tumorsize progress
XX348 proximal 6.3 0
XX987 proximal 10.0 0
> a[c(T,F,T),]
localisation tumorsize progress
XX348 proximal 6.3 0
XX987 proximal 10.0 0
> a$localisation
[1] "proximal" "distal" "proximal"
> a$localisation=="proximal"
[1] TRUE FALSE TRUE
> a[ a$localisation=="proximal", ]
localisation tumorsize progress
XX348 proximal 6.3 0
XX987 proximal 10.0 0
subset rows by a
vector of indices
subset rows by a
logical vector
subset a column
comparison resulting in
logical vector
subset the selected
rows
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-20-2048.jpg)


![• When the same or similar tasks need to be performed multiple
times for all elements of a list or for all columns of an array.
• May be easier and faster than “for” loops
• lapply(li, function )
• To each element of the list li, the function function is applied.
• The result is a list whose elements are the individual function
results.
> li = list("klaus","martin","georg")
> lapply(li, toupper)
> [[1]]
> [1] "KLAUS"
> [[2]]
> [1] "MARTIN"
> [[3]]
> [1] "GEORG"
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-29-2048.jpg)
![sapply( li, fct )
Like apply, but tries to simplify the result, by converting it into a
vector or array of appropriate size
> li = list("klaus","martin","georg")
> sapply(li, toupper)
[1] "KLAUS" "MARTIN" "GEORG"
> fct = function(x) { return(c(x, x*x, x*x*x)) }
> sapply(1:5, fct)
[,1] [,2] [,3] [,4] [,5]
[1,] 1 2 3 4 5
[2,] 1 4 9 16 25
[3,] 1 8 27 64 125
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-30-2048.jpg)
![apply( arr, margin, fct )
Apply the function fct along some dimensions of the array arr,
according to margin, and return a vector or array of the
appropriate size.
> x
[,1] [,2] [,3]
[1,] 5 7 0
[2,] 7 9 8
[3,] 4 6 7
[4,] 6 3 5
> apply(x, 1, sum)
[1] 12 24 17 14
> apply(x, 2, sum)
[1] 22 25 20
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-31-2048.jpg)

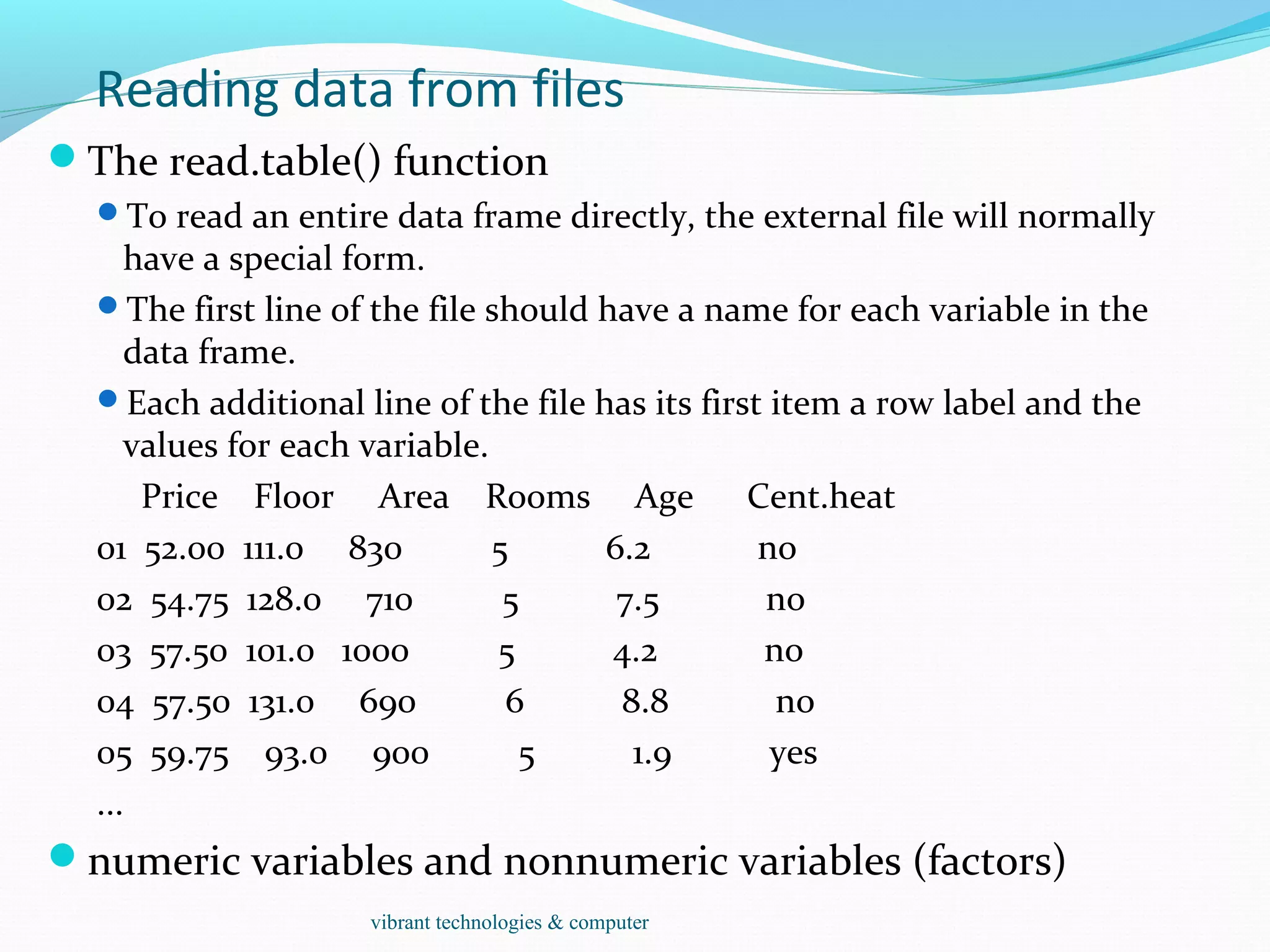
![Reading data from files
HousePrice <- read.table("houses.data", header=TRUE)
Price Floor Area Rooms Age Cent.heat
52.00 111.0 830 5 6.2 no
54.75 128.0 710 5 7.5 no
57.50 101.0 1000 5 4.2 no
57.50 131.0 690 6 8.8 no
59.75 93.0 900 5 1.9 yes
...
The data file is named ‘input.dat’.
Suppose the data vectors are of equal length and are to be read in in parallel.
Suppose that there are three vectors, the first of mode character and the
remaining two of mode numeric.
The scan() function
inp<- scan("input.dat", list("",0,0))
To separate the data items into three separate vectors, use assignments
like
label <- inp[[1]]; x <- inp[[2]]; y <- inp[[3]]
inp <- scan("input.dat", list(id="", x=0, y=0)); inp$id; inp$x; inp$y
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-35-2048.jpg)




![Implementation
How do we take a sample of residuals with replacement?
sample() is good for generating random samples of indices:
sample(10,rep=T) leads to “7 9 9 2 5 7 4 1 8 9”
Execute the bootstrap.
Make a matrix to save the results in and then repeat the bootstrap
process 1000 times for a linear regression with five regressors:
bcoef <- matrix(0,1000,6)
Program: for(i in 1:1000){
newy <- g$fit + g$res[sample(47, rep=T)]
brg <- lm(newy~y)
bcoef[i,] <- brg$coef
}
Here g is the output from the data with regression
analysis.
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-45-2048.jpg)
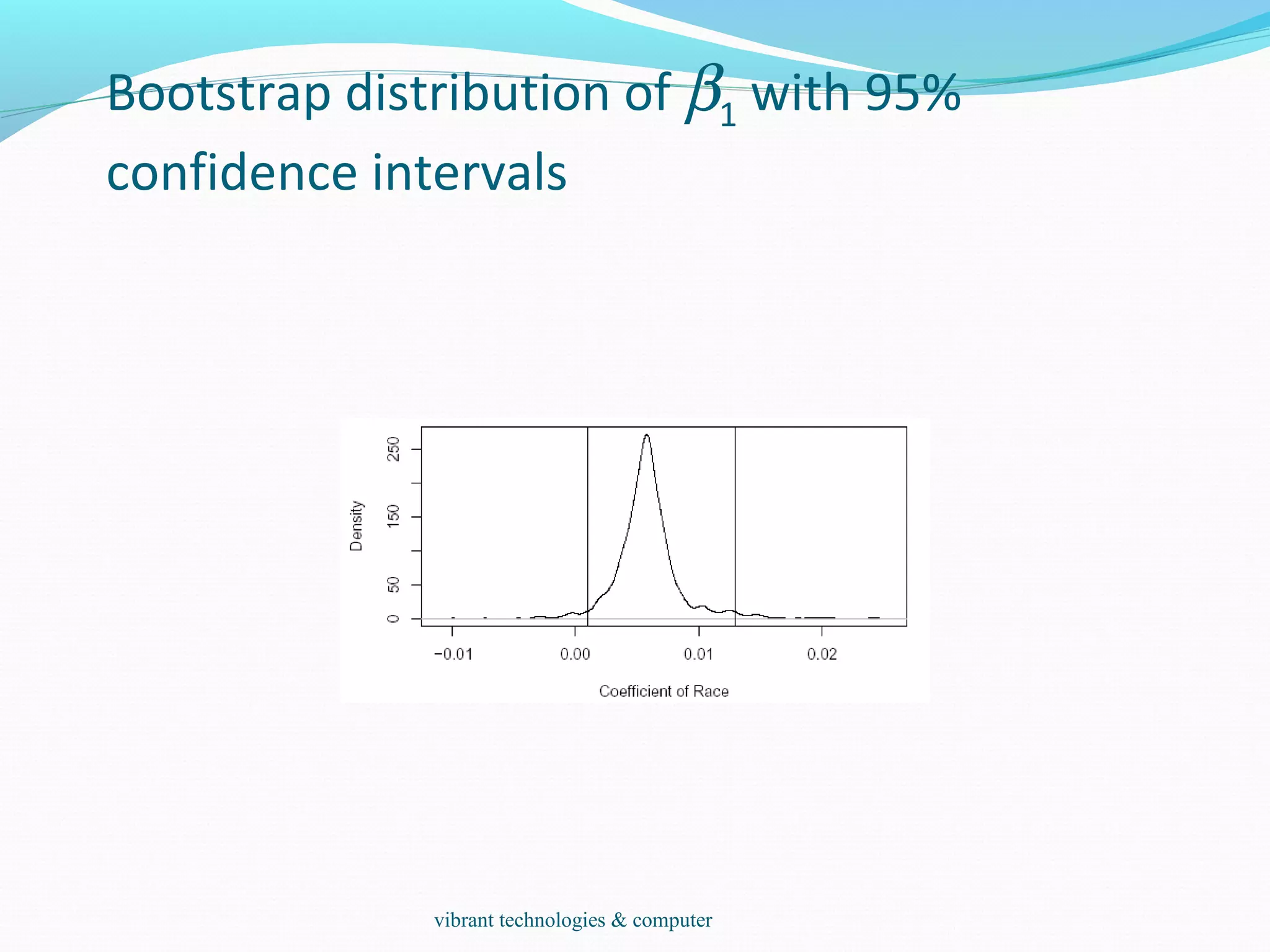
![Test and Confidence Interval
To test the null hypothesis that H0 : β1 = 0 against the
alternative H1 : β1 > 0, we may figure what fraction of the
bootstrap sampled β1 were less than zero:
length(bcoef[bcoef[,2]<0,2])/1000: It leads to 0.019.
The p-value is 1.9% and we reject the null at the 5% level.
We can also make a 95% confidence interval for this
parameter by taking the empirical quantiles:
quantile(bcoef[,2],c(0.025,0.975))
2.5% 97.5%
0.00099037 0.01292449
We can get a better picture of the distribution by looking at
the density and marking the confidence interval:
plot(density(bcoef[,2]),xlab="Coefficient of Race",main="")
abline(v=quantile(bcoef[,2],c(0.025,0.975)))
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-46-2048.jpg)
![The Effect of Choice of Kernels
The average amount of annual precipitation (rainfall) in
inches for each of 70 United States (and Puerto Rico) cities.
data(precip)
bw <- bw.SJ(precip) ## sensible automatic choice
plot(density(precip, bw = bw, n = 2^13), main = "same sd
bandwidths, 7 different kernels")
for(i in 2:length(kernels)) lines(density(precip, bw = bw, kern
= kernels[i], n = 2^13), col = i)
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-48-2048.jpg)
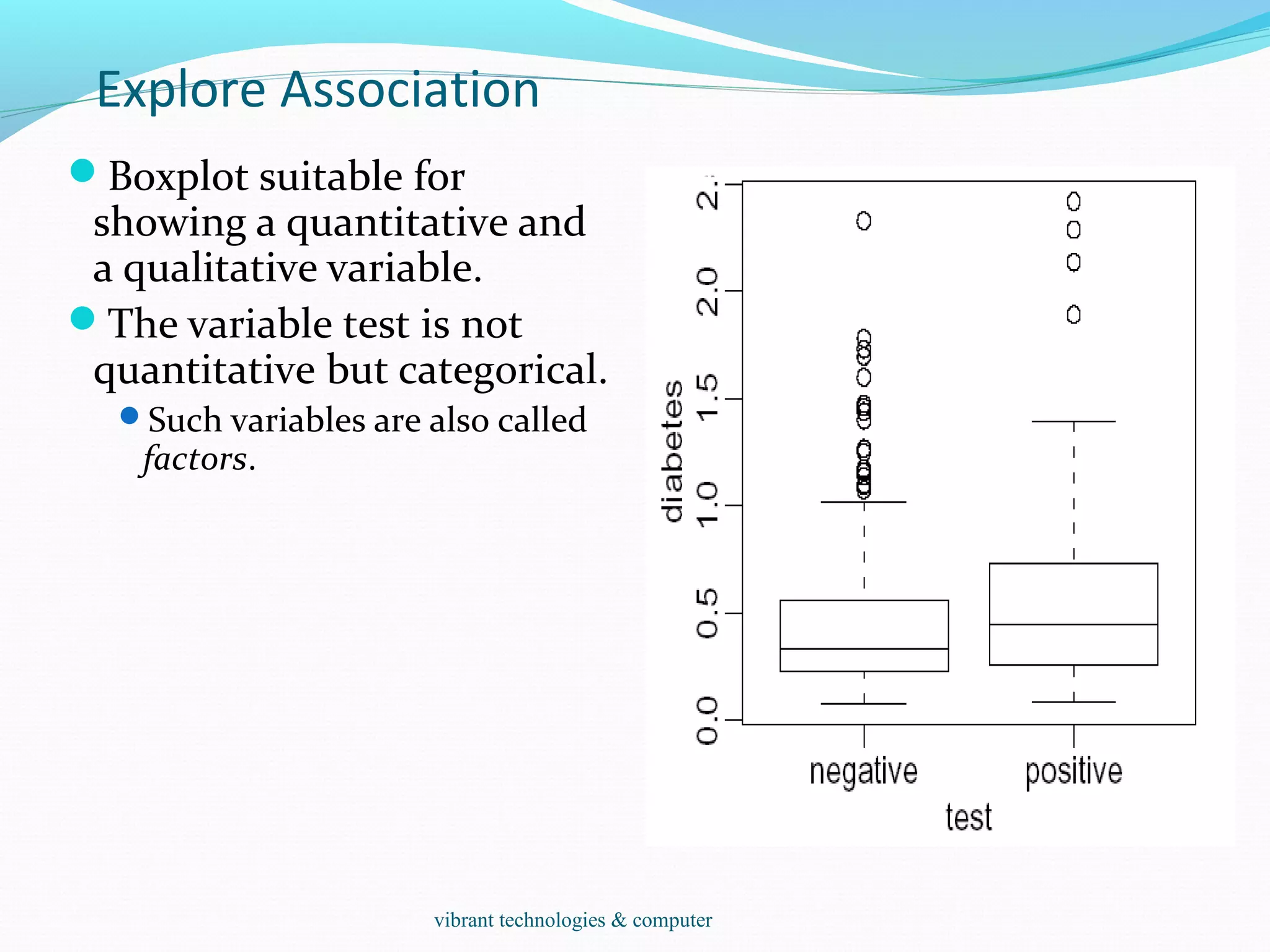
![Explore Association
Data(stackloss)
It is a data frame with 21 observations on 4 variables.
[,1] `Air Flow' Flow of cooling air
[,2] `Water Temp' Cooling Water Inlet Temperature
[,3] `Acid Conc.' Concentration of acid [per 1000, minus
500]
[,4] `stack.loss' Stack loss
The data sets `stack.x', a matrix with the first three
(independent) variables of the data frame, and `stack.loss',
the numeric vector giving the fourth (dependent) variable,
are provided as well.
Scatterplots, scatterplot matrix:
plot(stackloss$Ai,stackloss$W)
plot(stackloss) data(stackloss)
two quantitative variables.
summary(lm.stack <- lm(stack.loss ~ stack.x))
summary(lm.stack <- lm(stack.loss ~ stack.x))
vibrant technologies & computer](https://image.slidesharecdn.com/r-programming-training-in-mumbai-150916114531-lva1-app6891/75/R-programming-training-in-mumbai-49-2048.jpg)