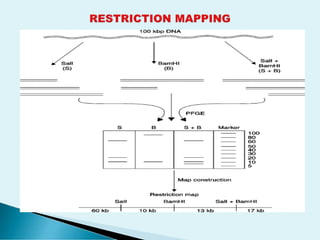
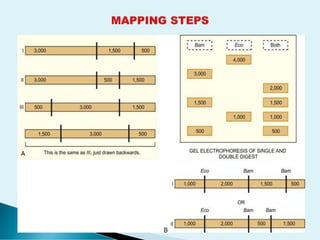
Genetic Engineering and Biotechnology is not a mere a fancy science but it is the call of the time to think biology differently for the sustainability in all respects. The restricted endonuclease or popularly called molecular sessiors are required to develop the science for its multiple applications.The restricted endonuclease not only cut the DNA like sessiors but also it enables to find out the different molecular length in terms of base pairs for the exact precision. The presentation offers a bird's eye view about the different type of restricted endovnucleases and how they are being employed to explore the molecular beauty of plasmid or linear DNA molecule along with their importance.