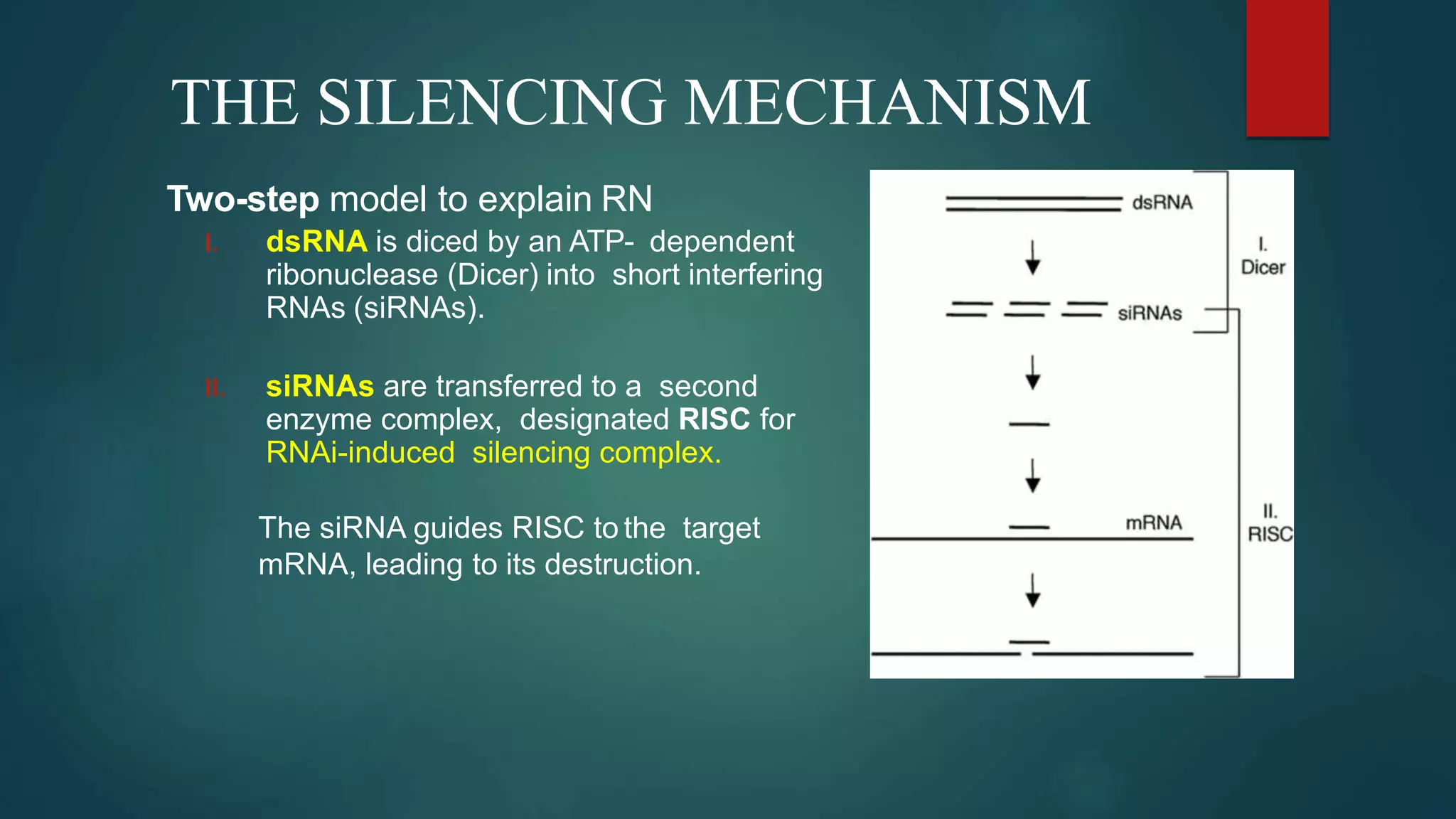
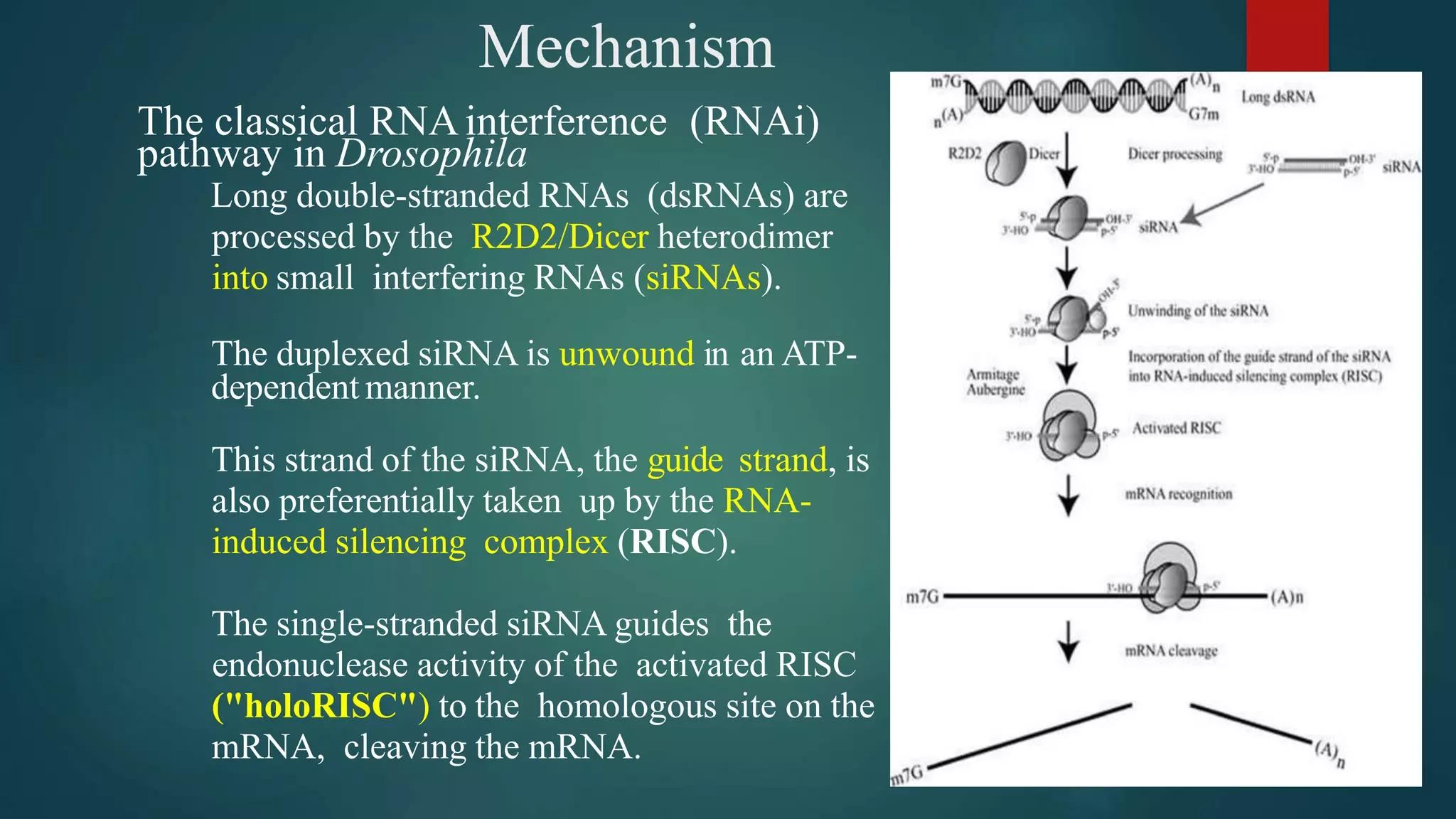
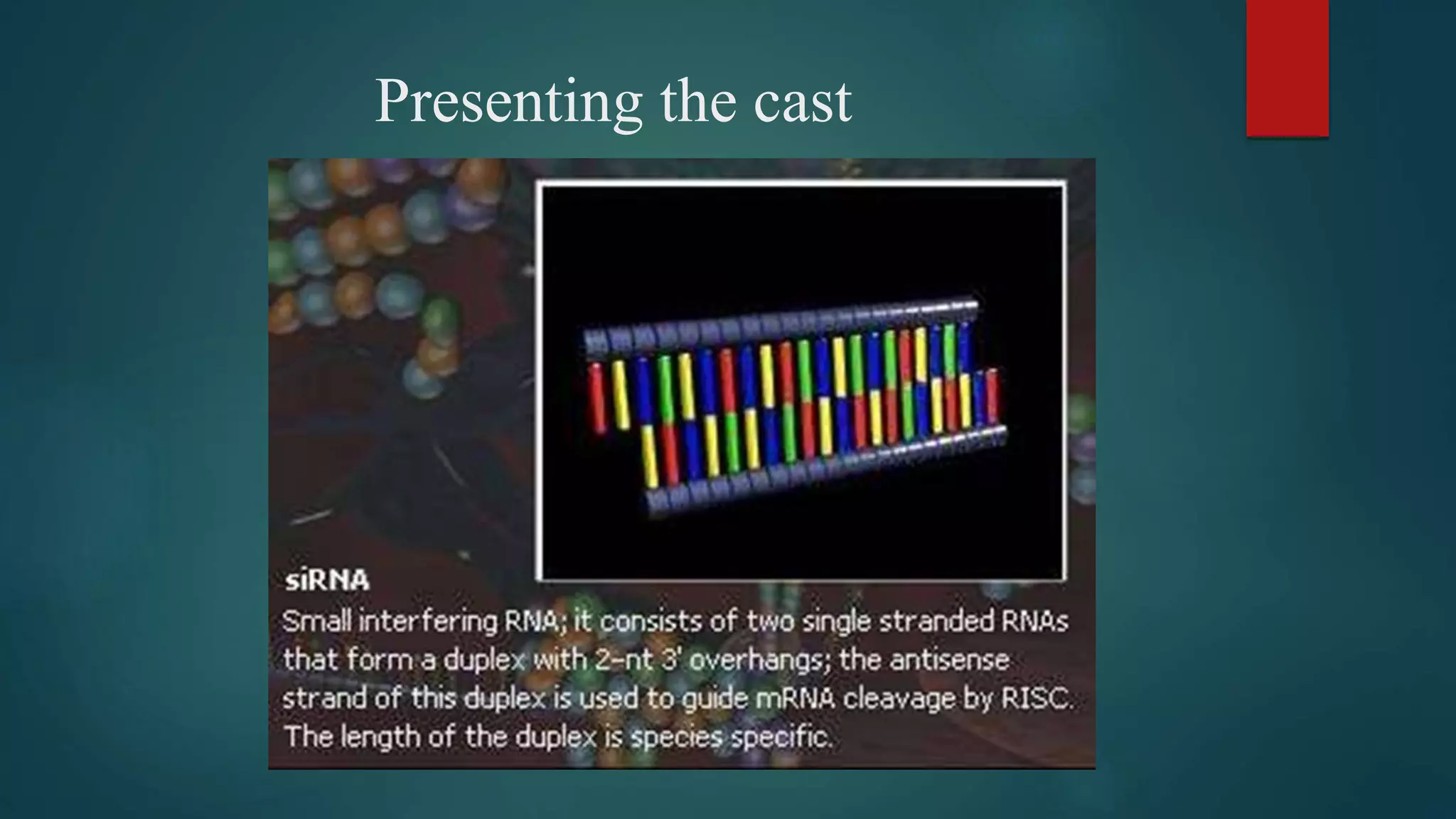
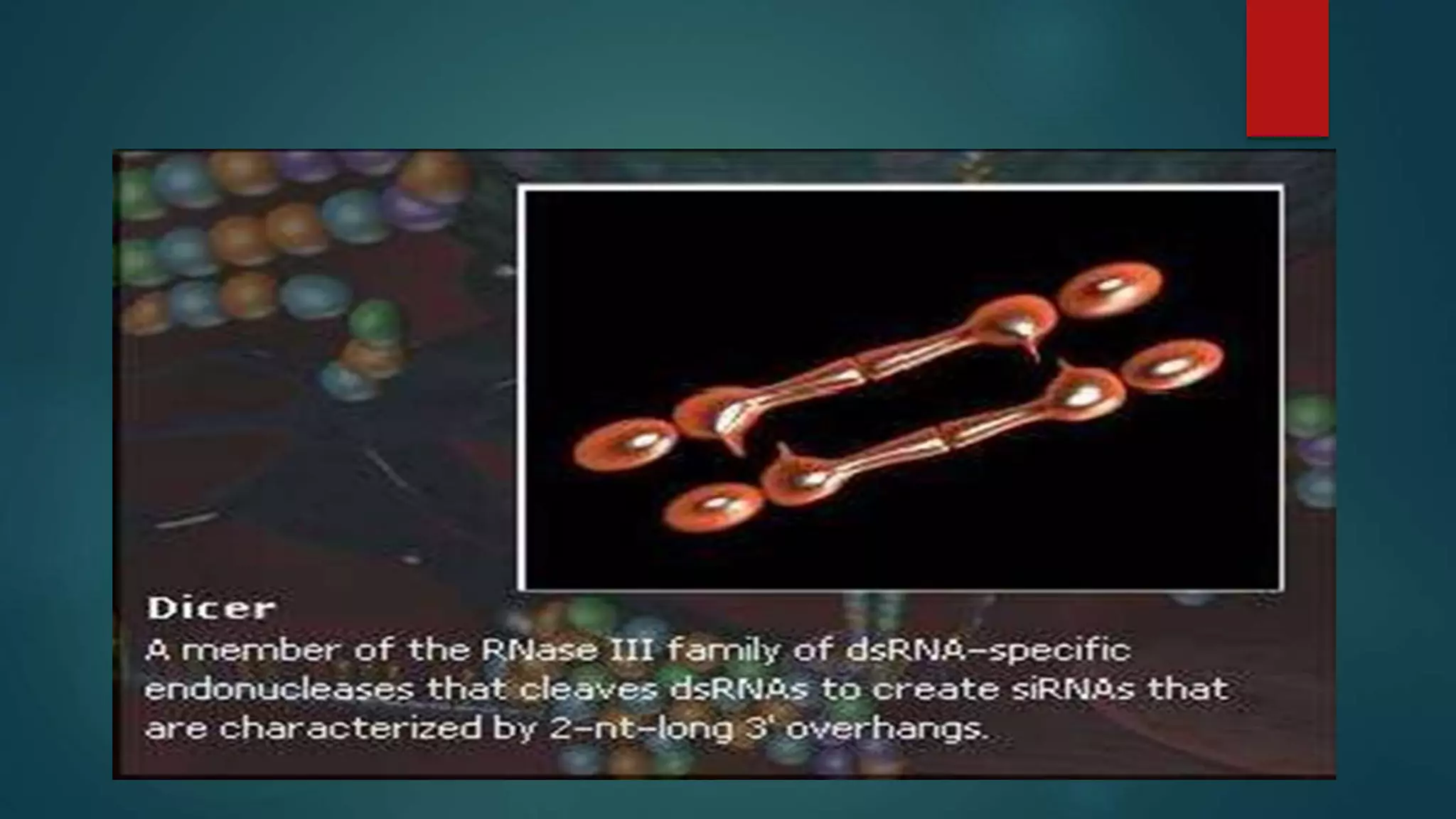
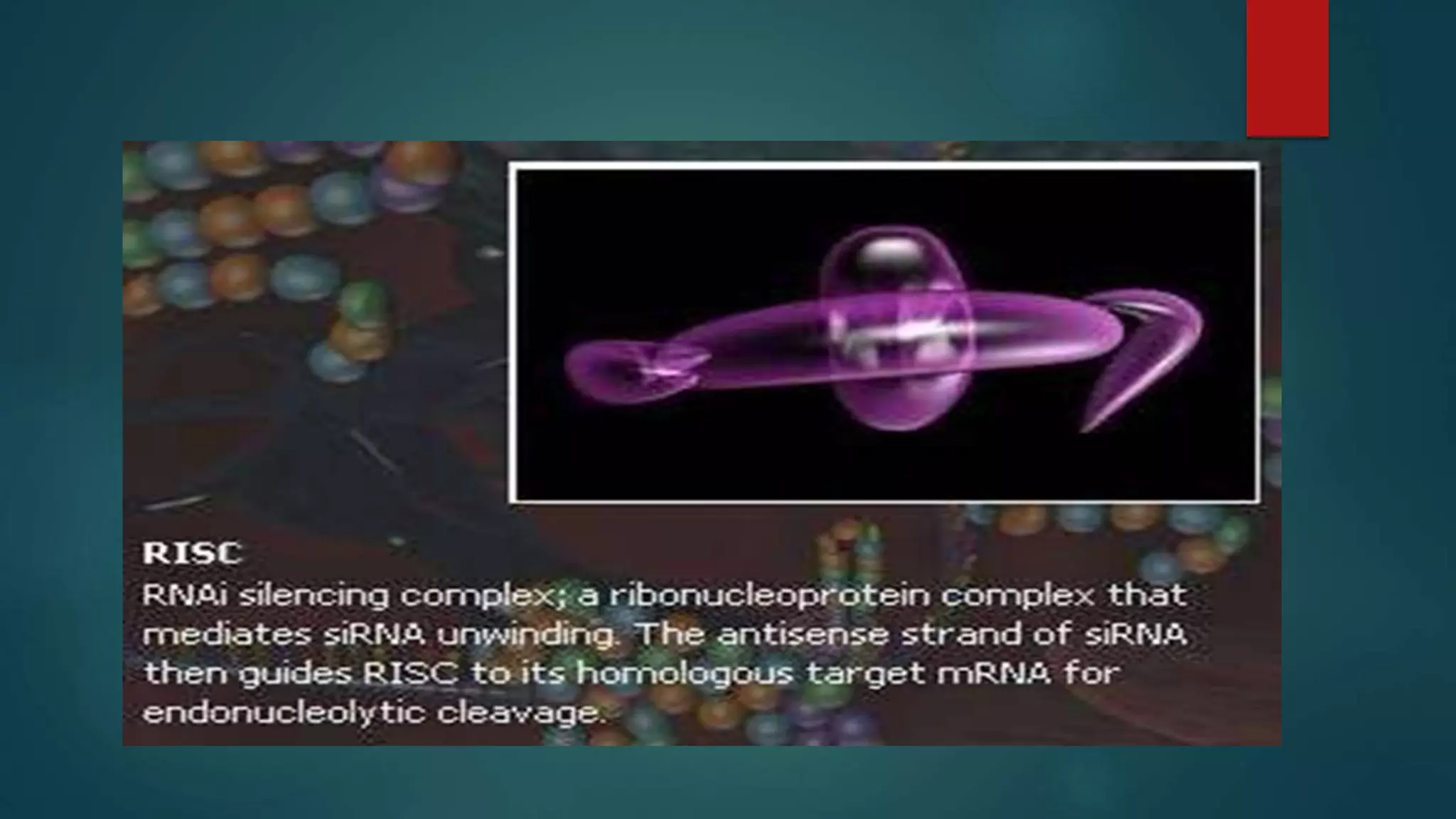
RNA interference (RNAi) is a conserved biological process that silences gene expression via the degradation of specific mRNA through small double-stranded RNA (siRNA) molecules, first discovered in 1998. The mechanism involves Dicer enzyme processing long dsRNA into 21-23 nt siRNA, which then directs the RNA-induced silencing complex (RISC) to cleave homologous mRNA strands. This silencing mechanism is vital for cellular defense against viral invasion and is seen across various eukaryotic organisms.