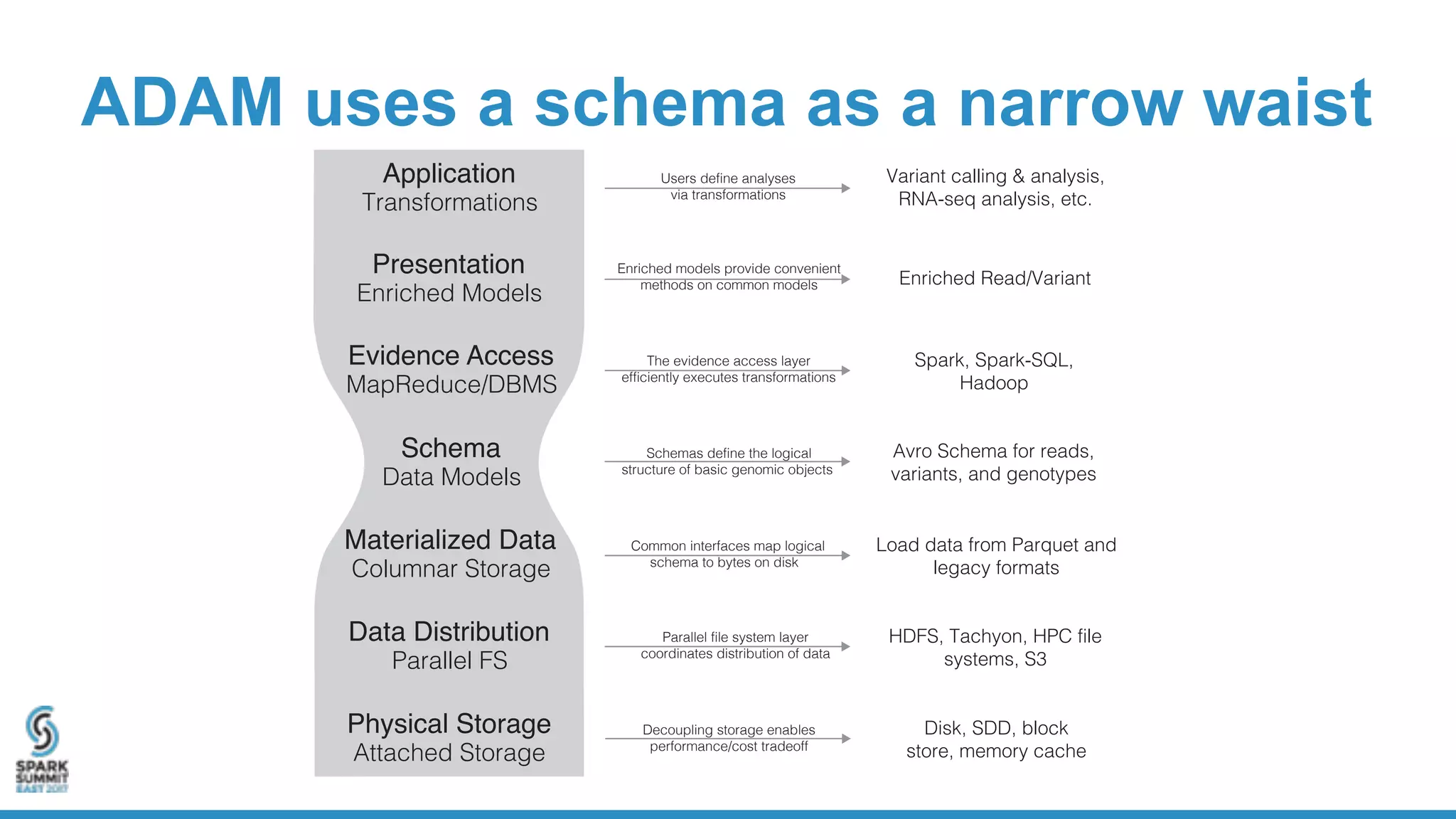
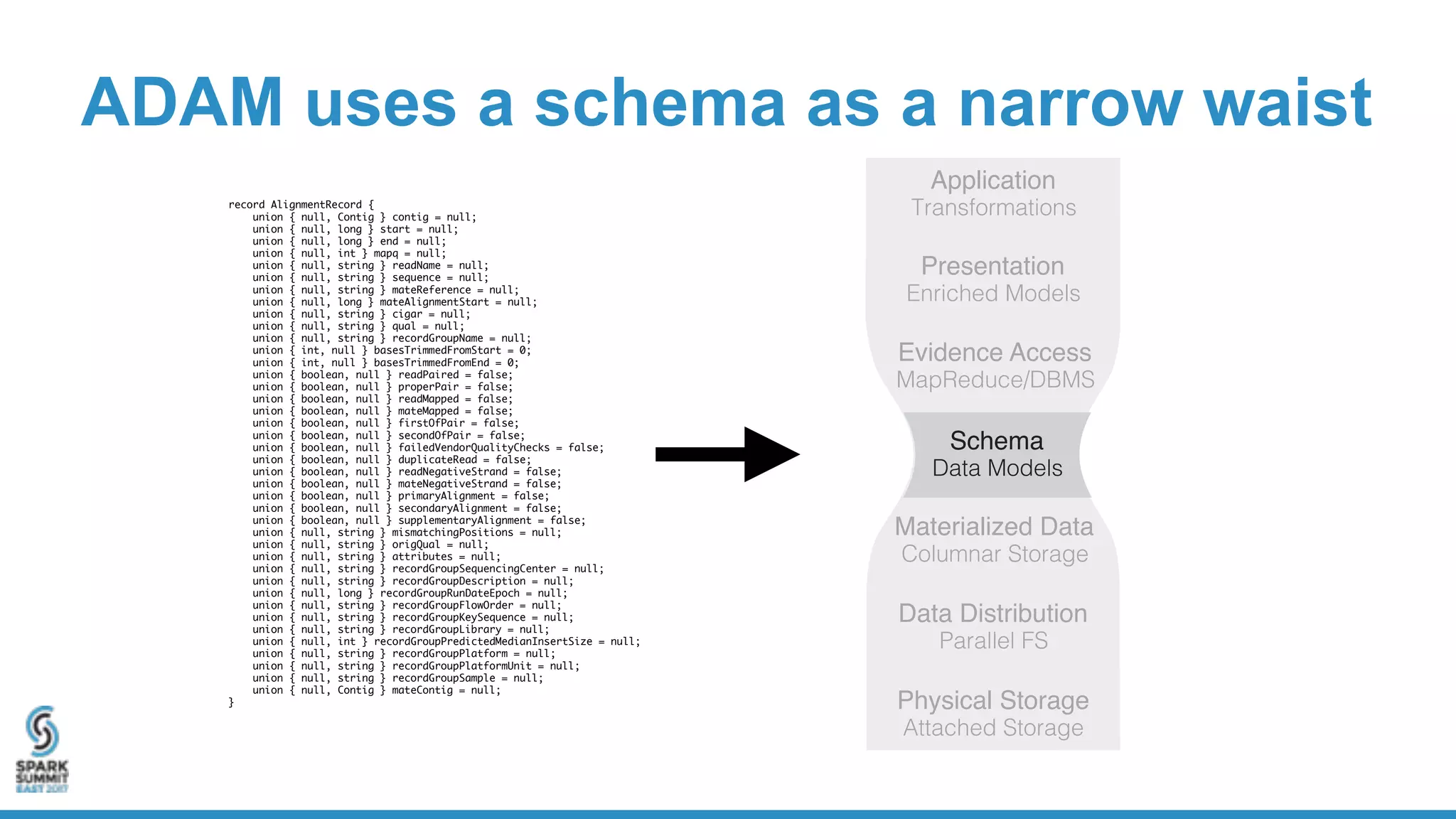
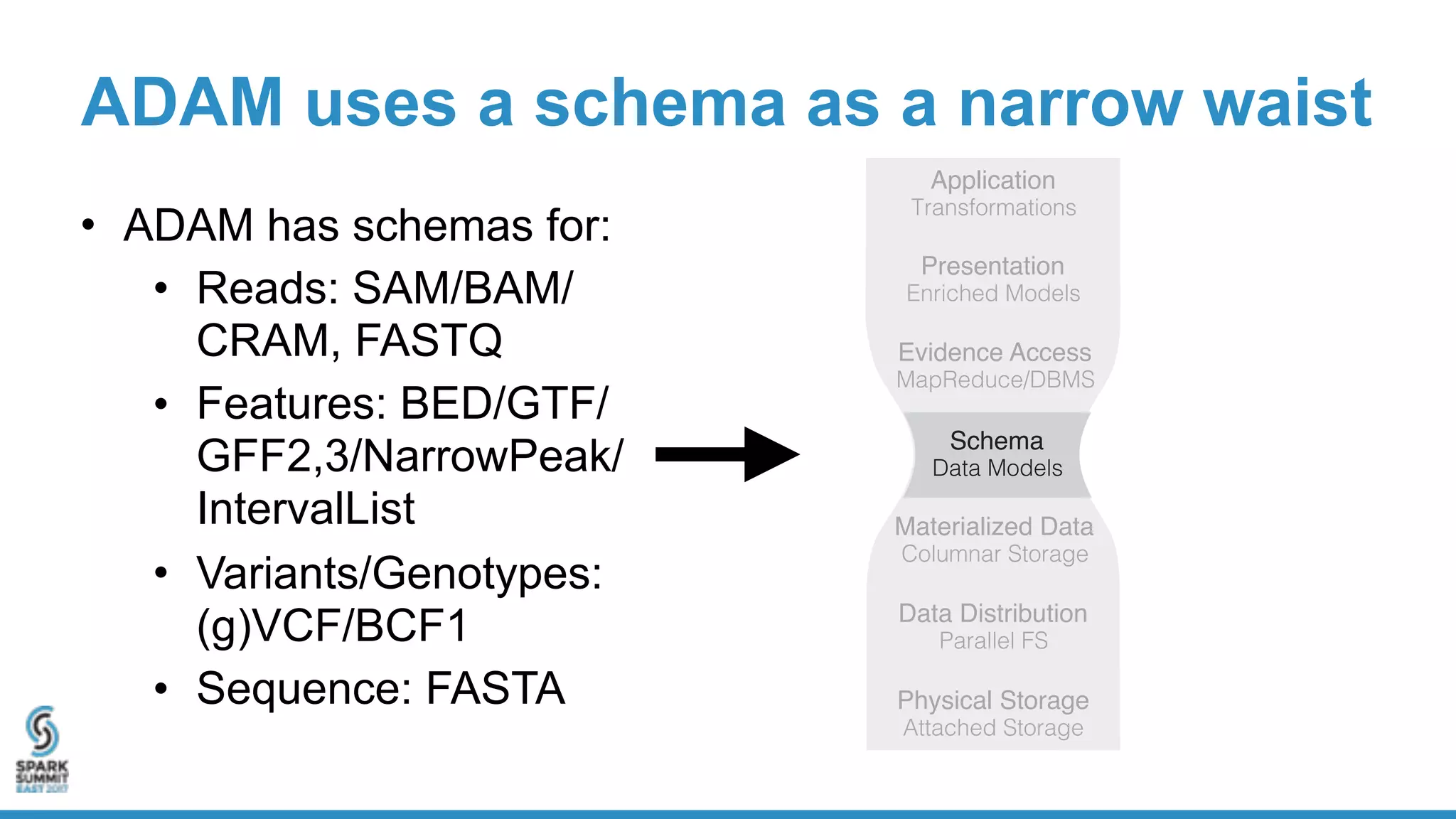
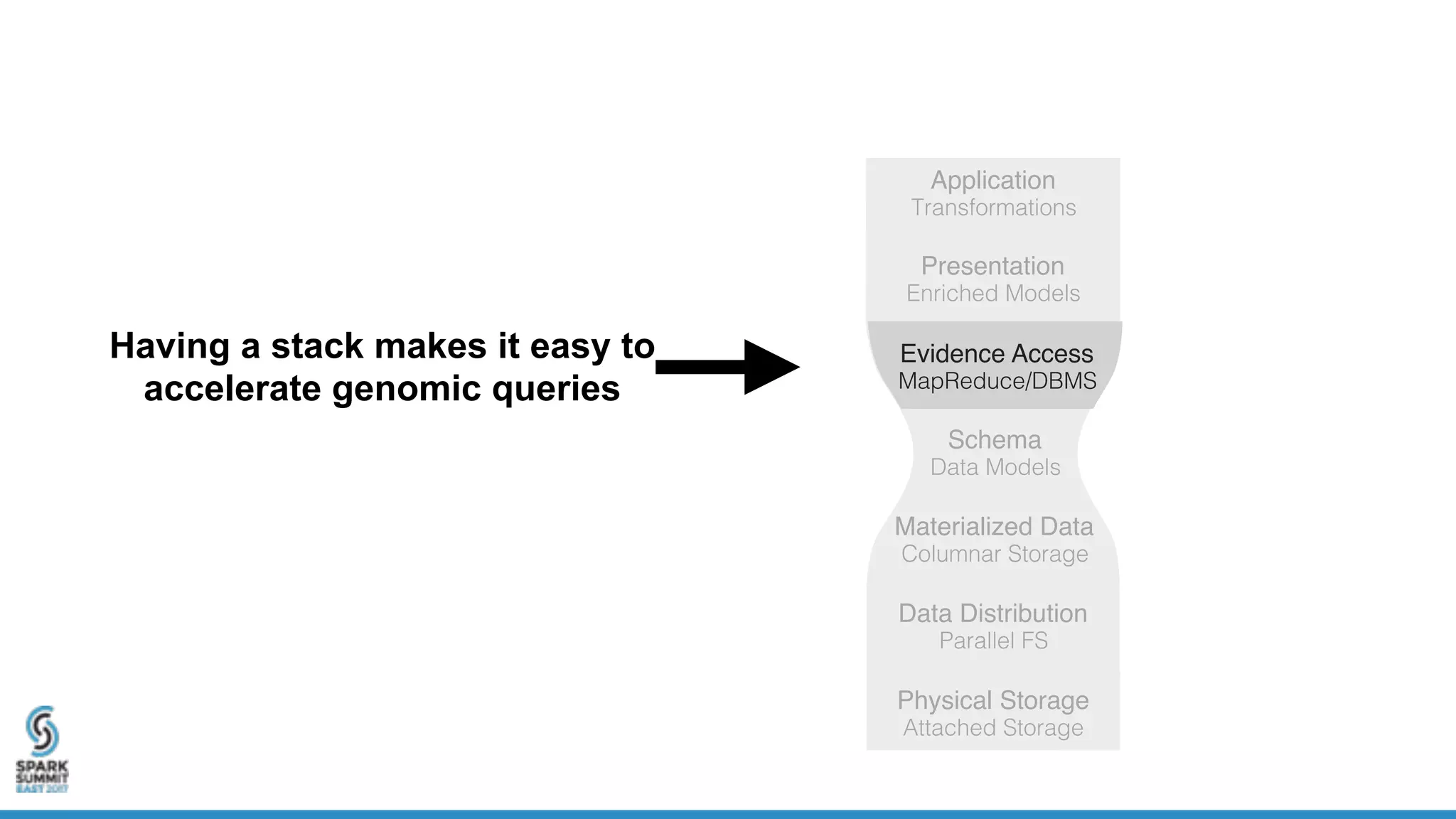
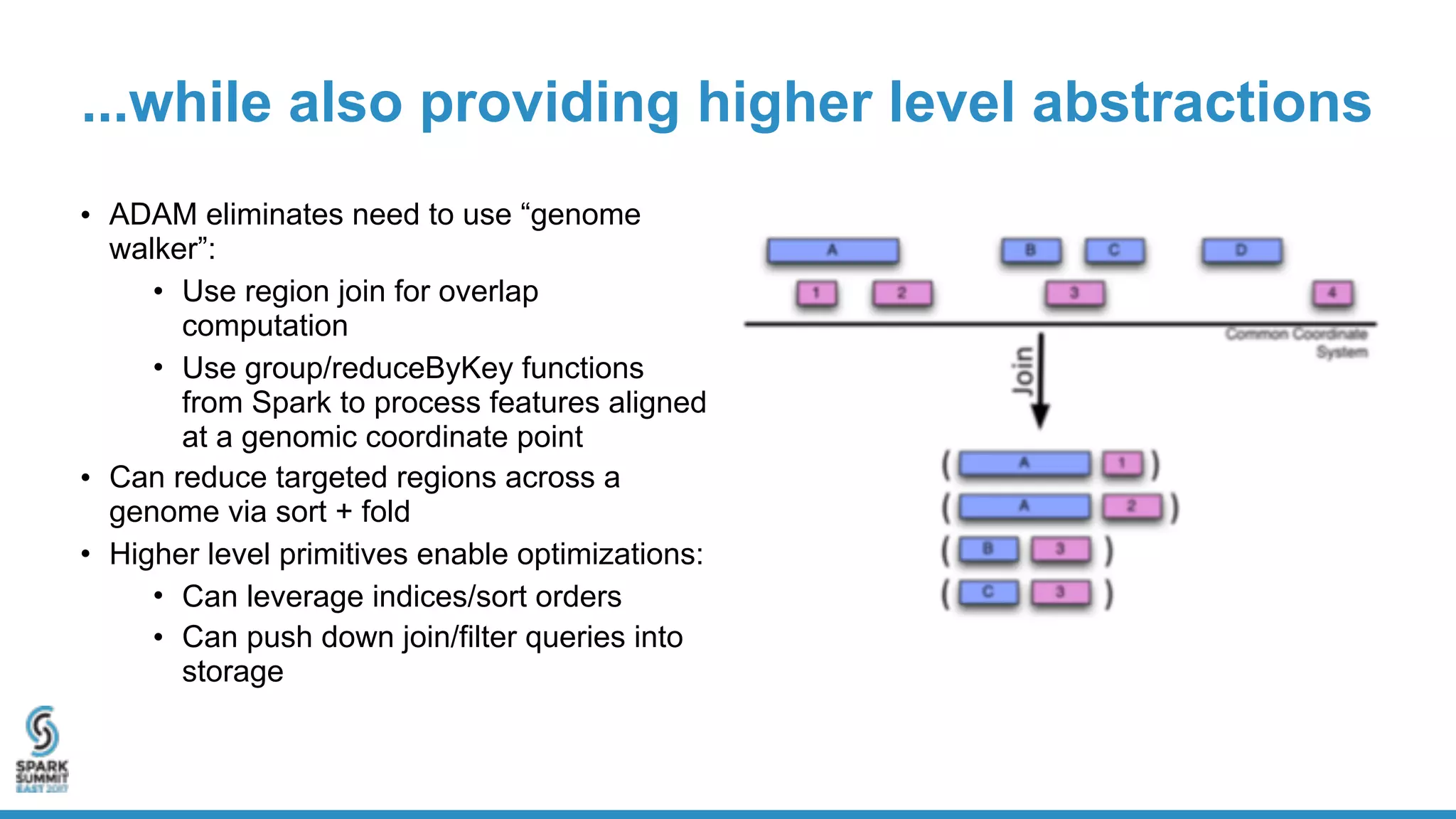
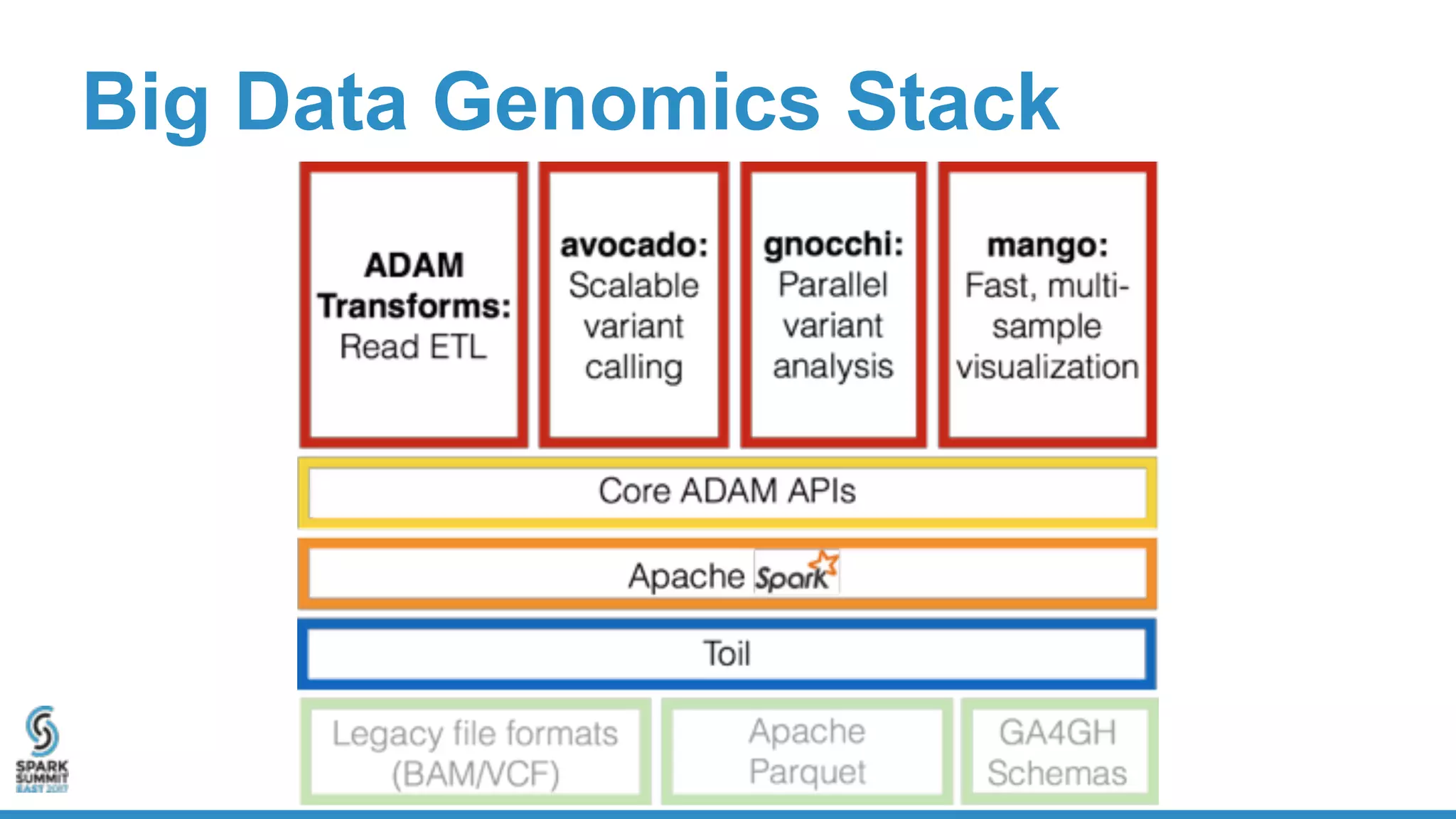
- ADAM is an open source, high performance, distributed library for genomic analysis that uses Spark and Scala. It defines schemas and interfaces for processing and analyzing large genomic datasets in a distributed manner.
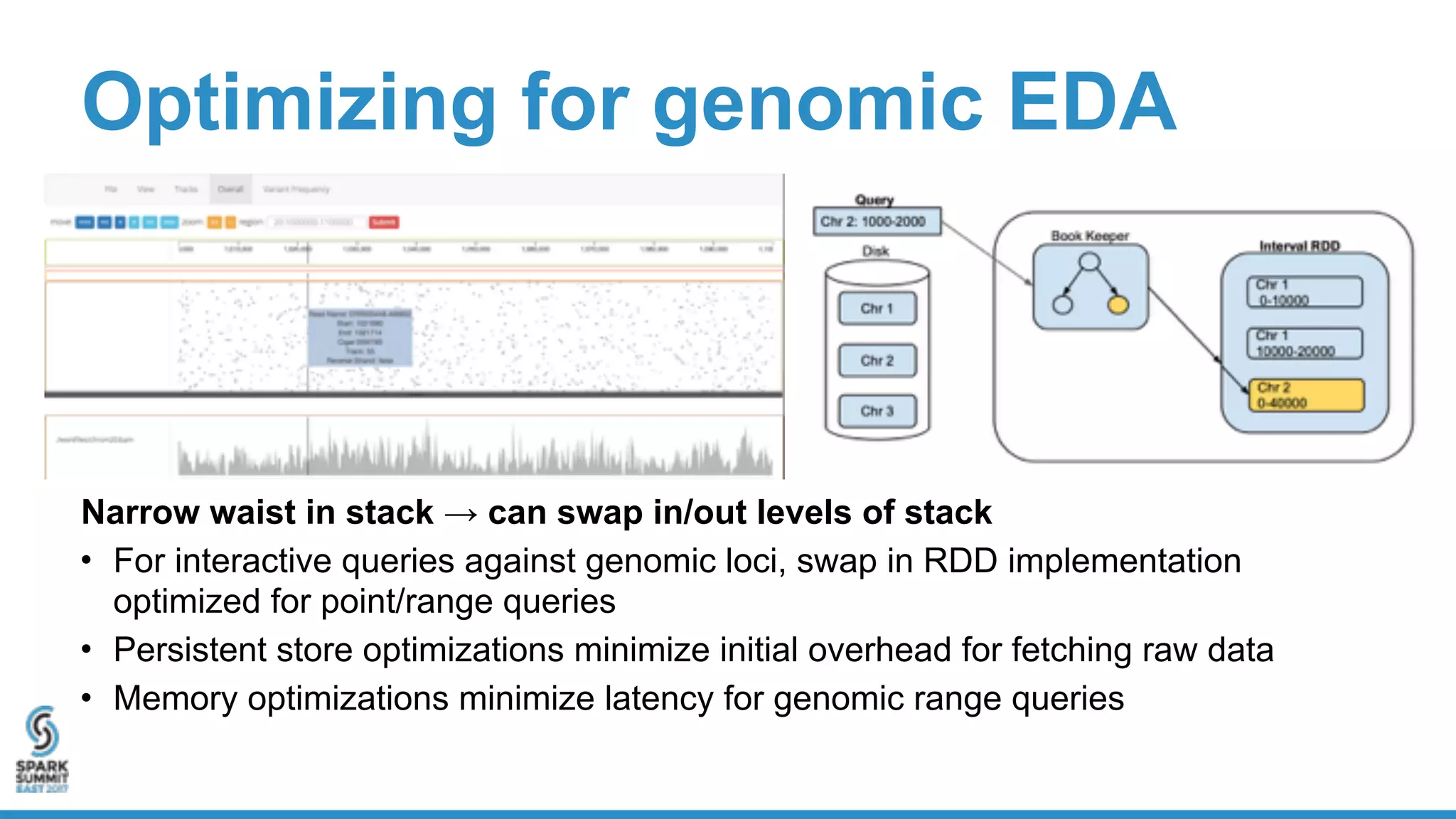
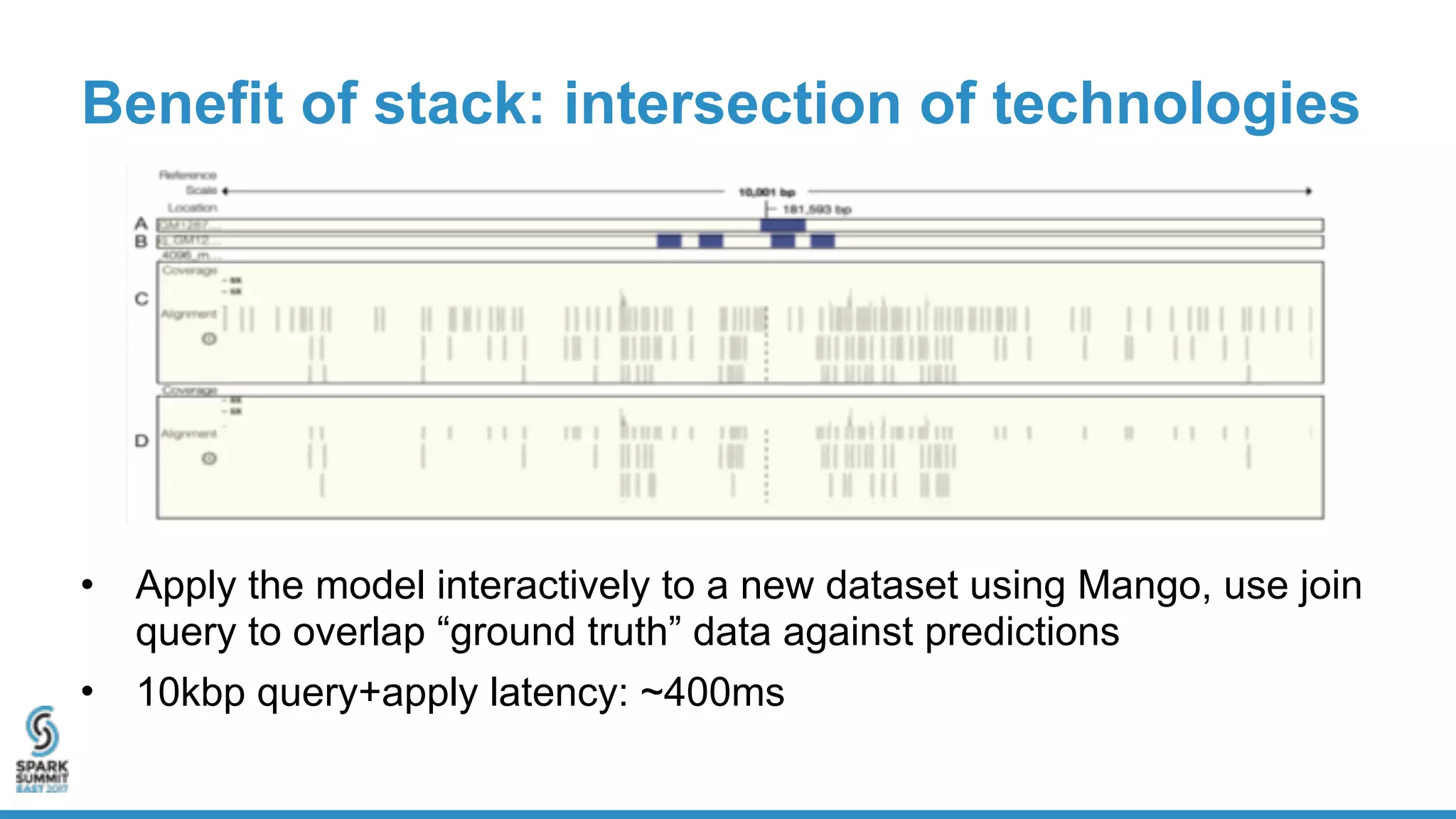
- ADAM provides schemas for common genomic file formats like SAM/BAM, VCF, FASTA. It enables both batch and interactive analysis of genomic data while optimizing for performance and avoiding data lock-in issues.
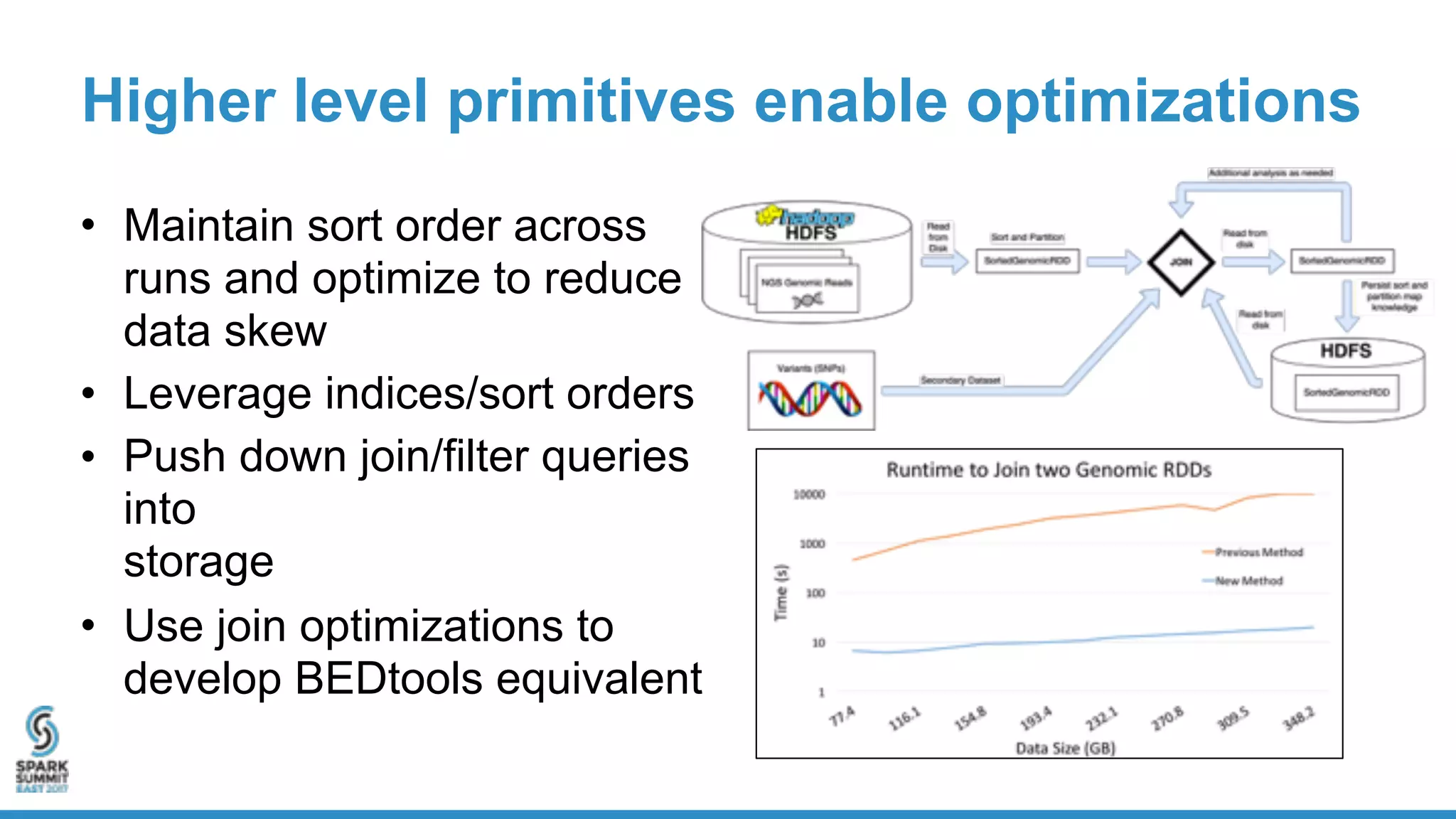
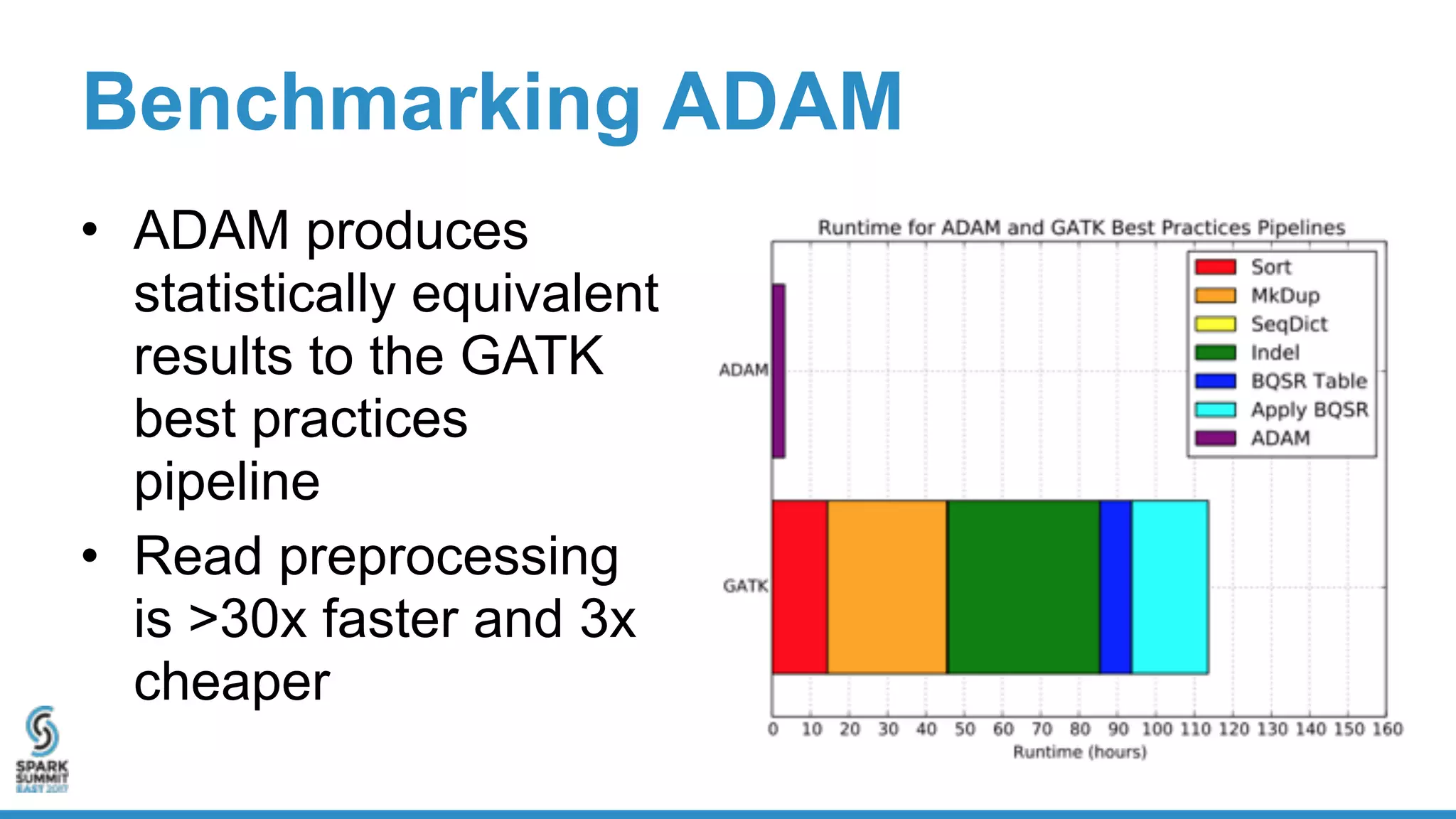
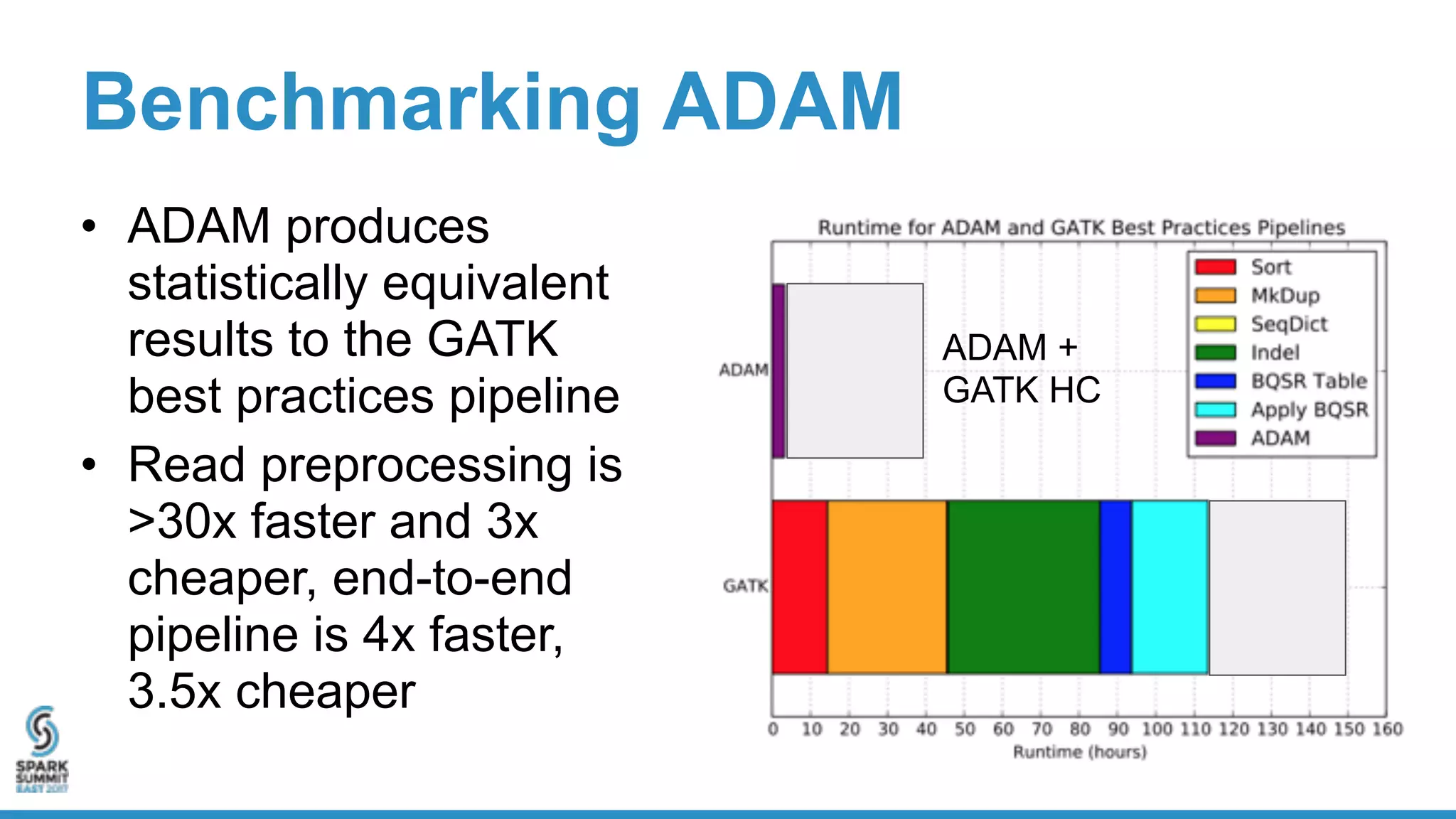
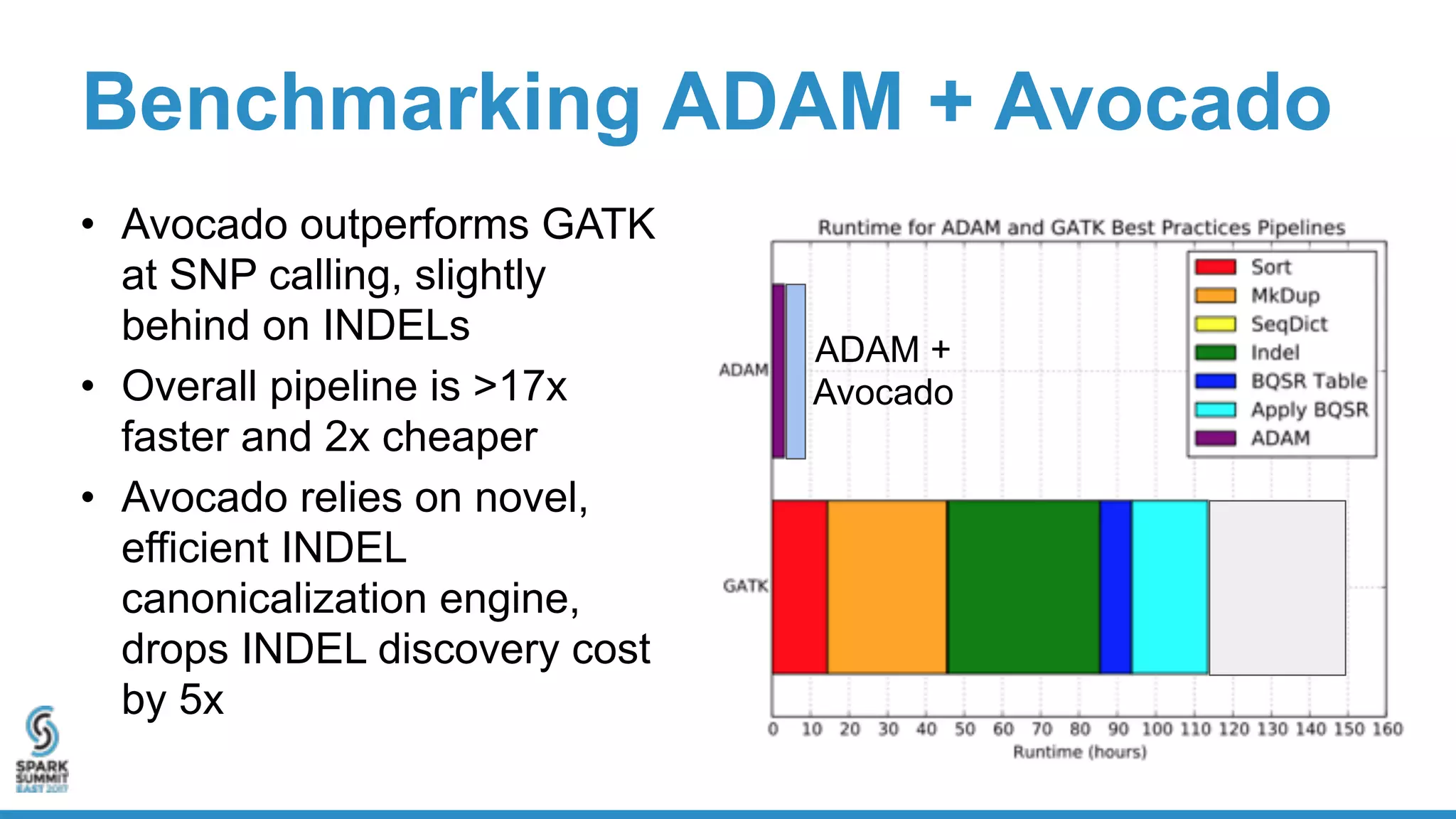
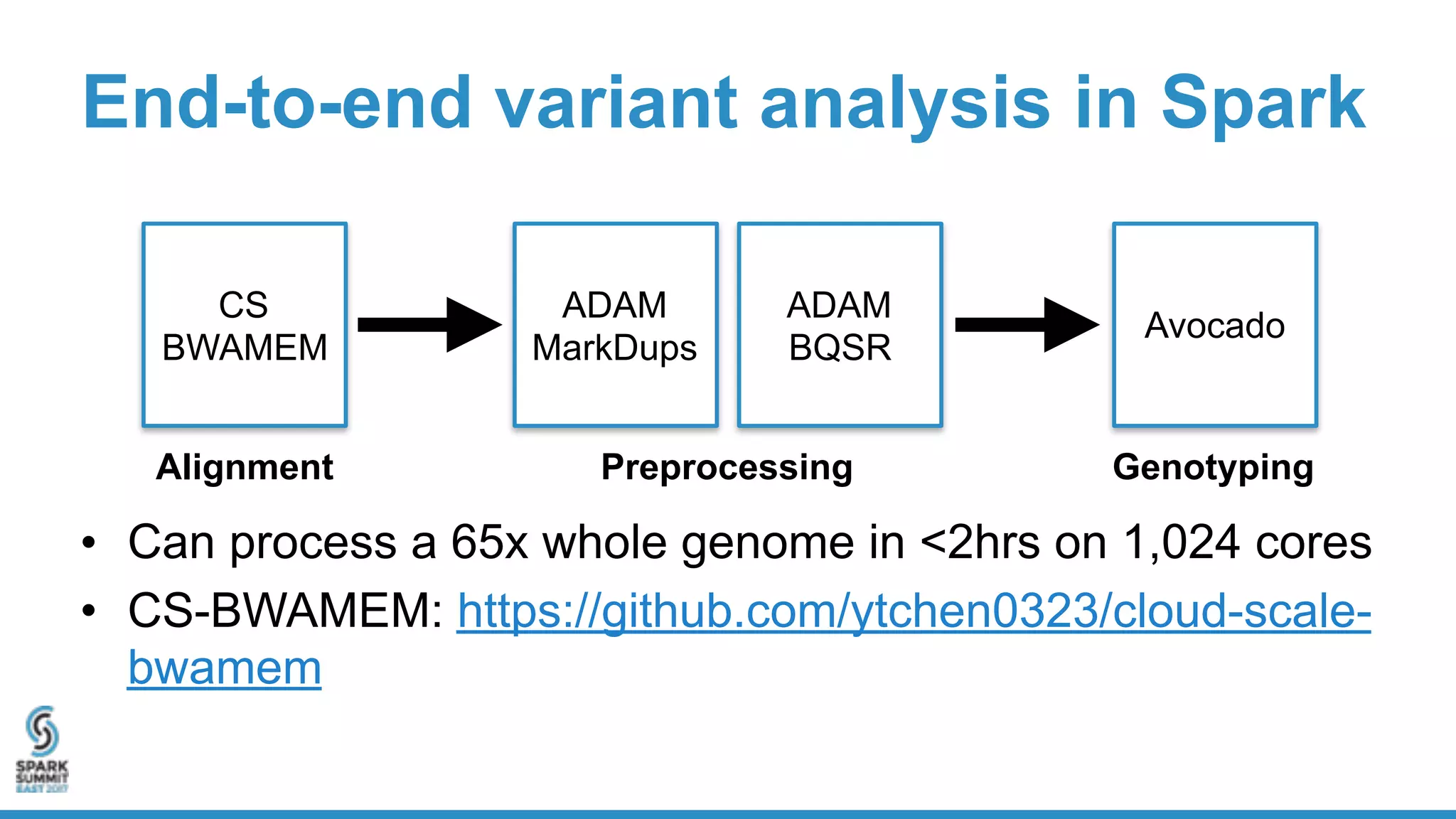
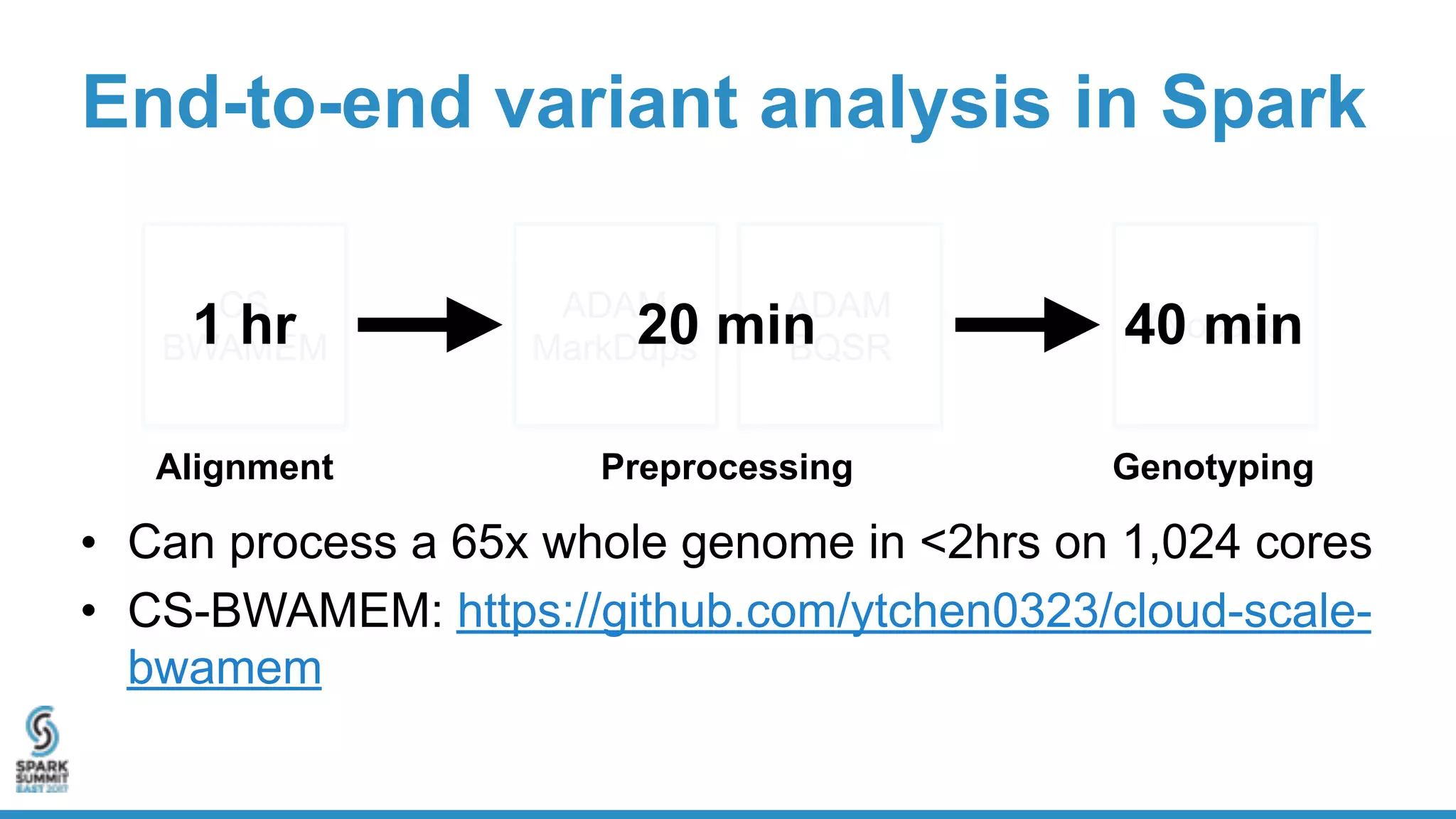
- Benchmarking shows ADAM can process a 65x human genome dataset faster and cheaper than existing tools, enabling end-to-end analysis in under 2 hours on 1,024 cores.