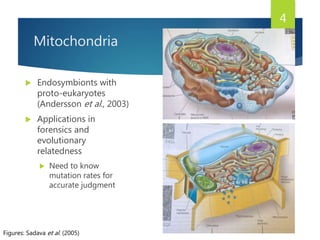
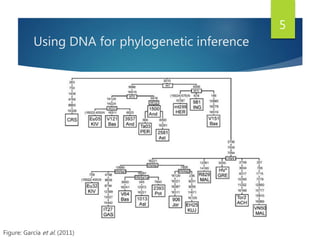
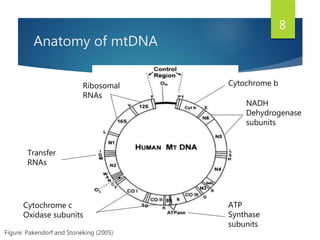
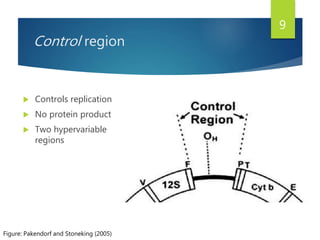
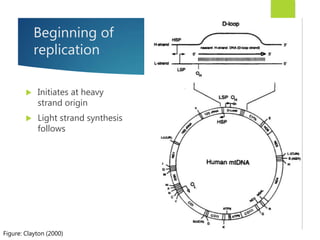
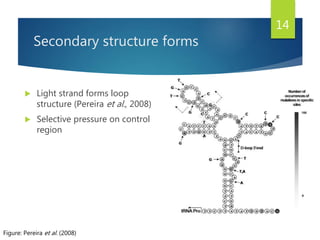
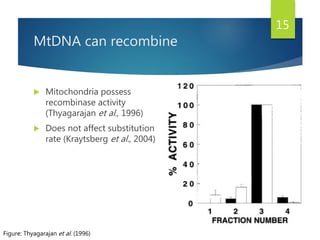
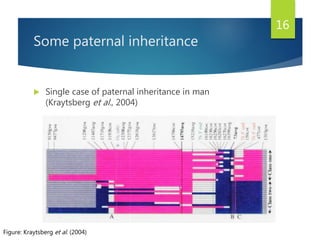
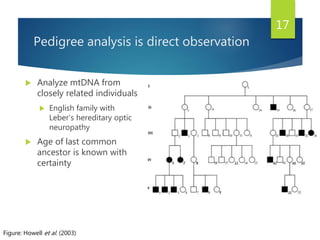
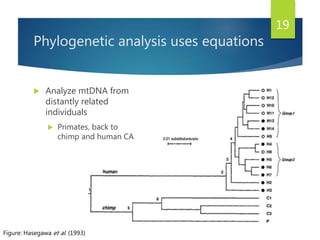
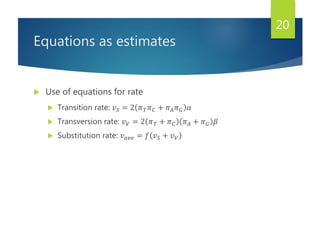
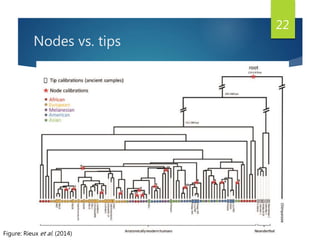
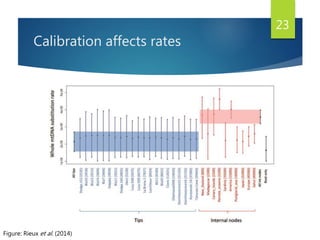
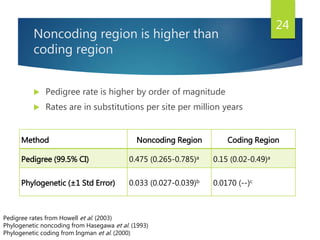
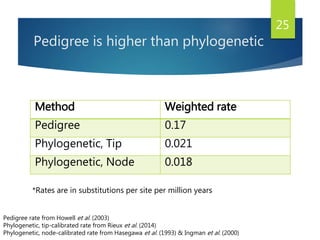
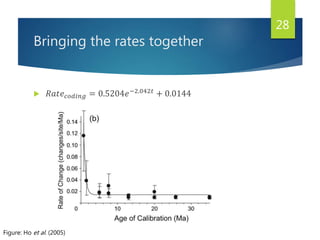
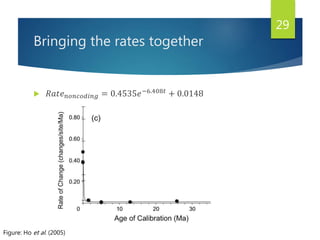
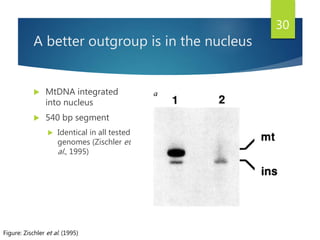
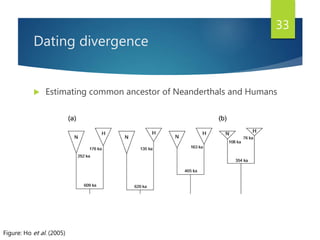
This document discusses methods for analyzing the human mitochondrial DNA (mtDNA) substitution rate. It describes three main methods: pedigree analysis of closely related individuals, phylogenetic analysis of distantly related individuals using equations, and bringing the rates together using an exponential equation. Pedigree analysis tends to overestimate the rate while phylogenetic analysis may underestimate due to uncertainty in time. The document also discusses improving current findings by using a better nuclear DNA outgroup and looking at coding versus noncoding substitution rates. Understanding mtDNA substitution rates has applications in forensics, dating species divergence, and determining relatedness to ancestors over time.