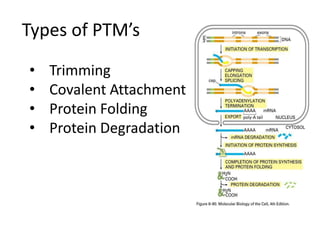
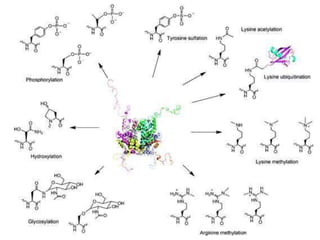
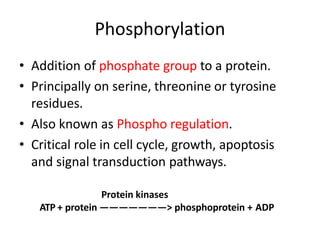
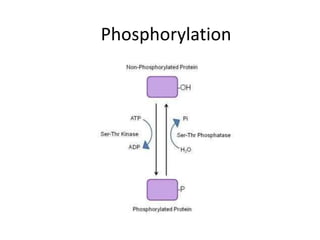
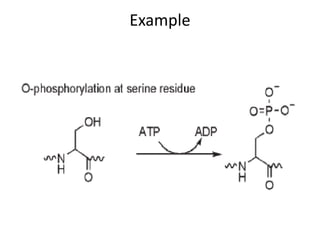
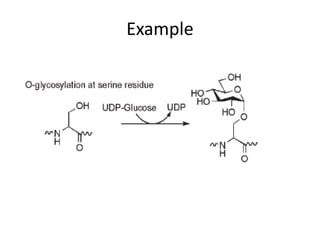
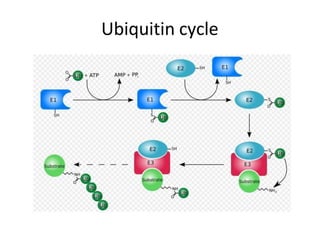
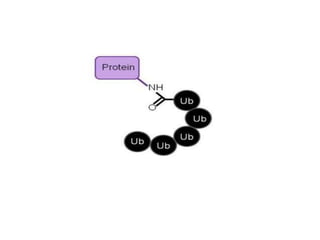
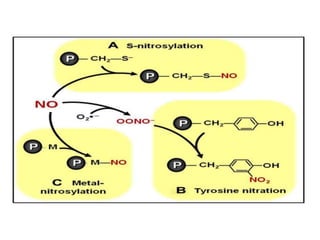
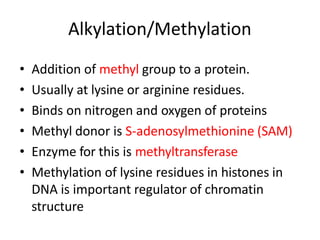
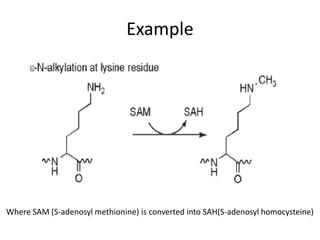
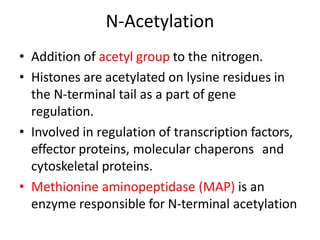
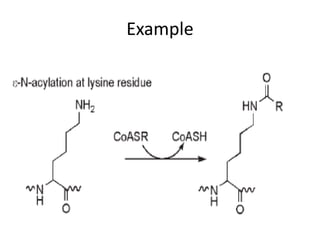
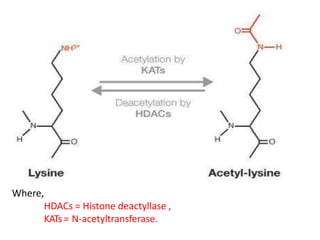
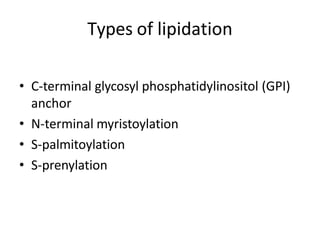
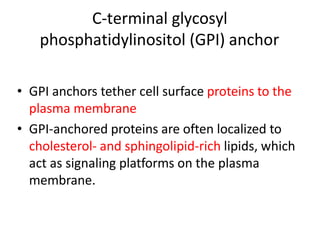
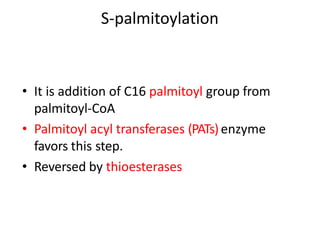
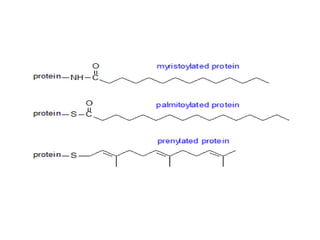
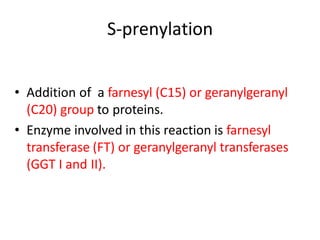
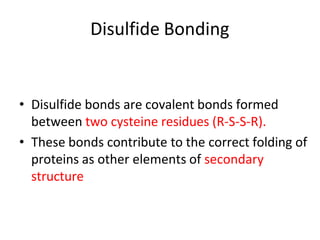
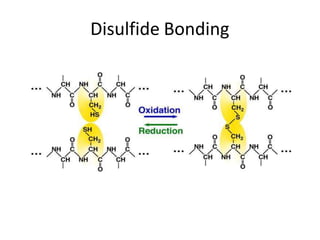
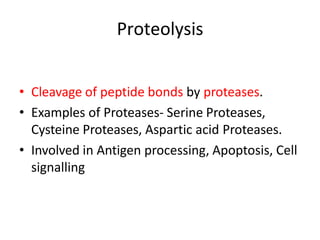
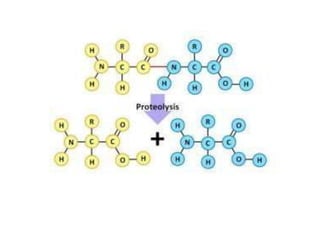
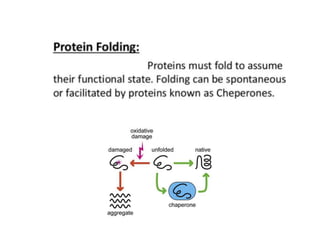
Post-translational modifications (PTMs) are chemical changes that occur to proteins after translation. PTMs regulate protein activity, localization, and interactions. The main types of PTMs are phosphorylation, glycosylation, ubiquitination, and methylation. Phosphorylation involves the addition of phosphate groups and is important for cell signaling. Glycosylation adds carbohydrate groups and affects protein structure. Ubiquitination tags proteins for destruction, and methylation adds methyl groups, regulating processes like gene expression. PTMs are identified through techniques like mass spectrometry and chromatographic analysis.