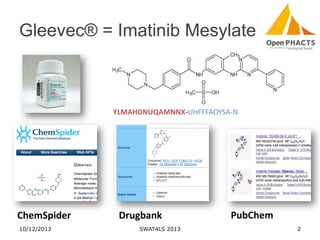
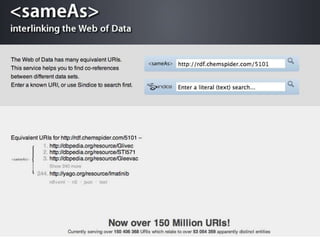
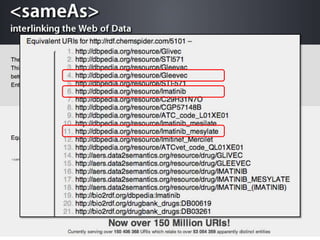
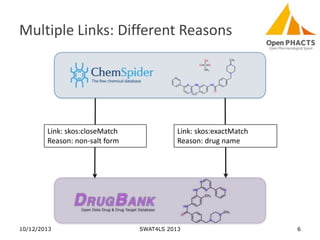
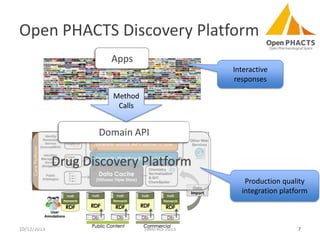
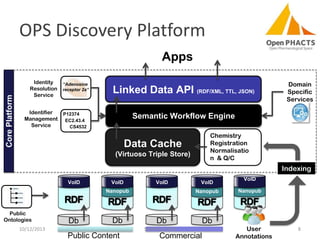
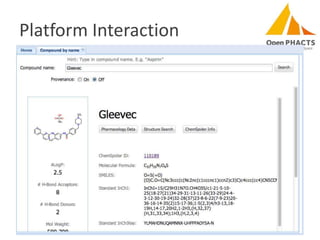
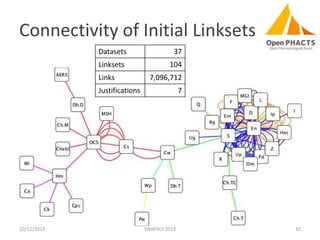
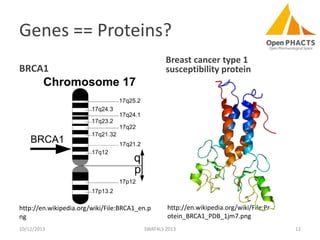
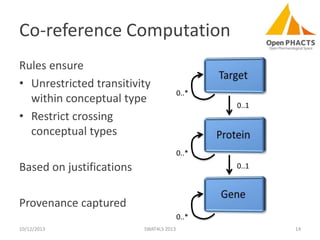
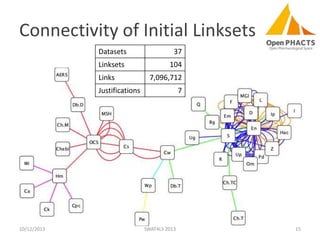
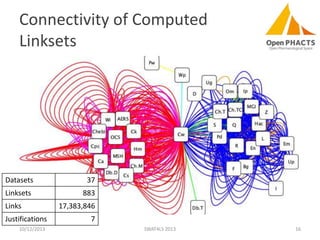
The document discusses the challenges of identity co-reference in drug discovery datasets, emphasizing the concept that the number of identifiers for an individual may equal or exceed the number of institutions involved. It outlines a method for resolving identities, including the implementation of rules to ensure accurate connections between genes, proteins, and compounds, while capturing provenance. Conclusions highlight the advantages of computing co-reference, which includes reduced data requirements and increased dataset coverage.