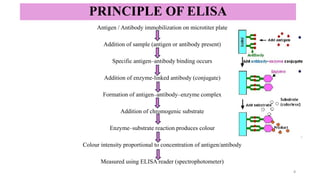
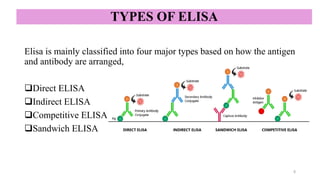
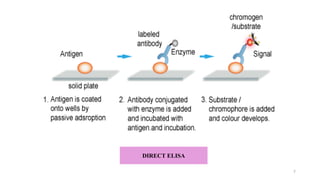
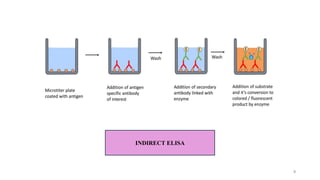
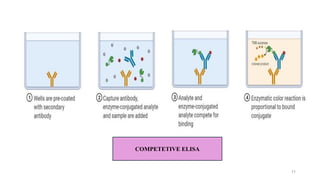
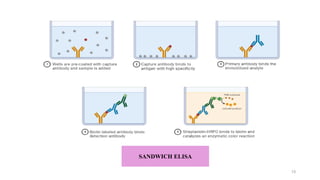
ELISA-a sensitive and specific technique used for the detection and
quantification of antigens or antibodies in biological samples.
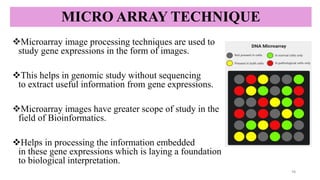
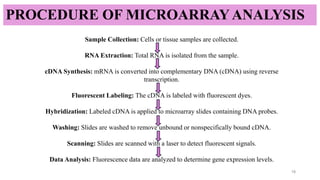
Microarray technique-enables the simultaneous analysis of thousands of
genes to study gene expression and genetic variations efficiently.
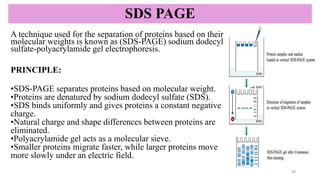
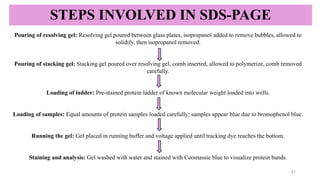
SDS-PAGE-a reliable technique used to separate proteins based on their
molecular weight.
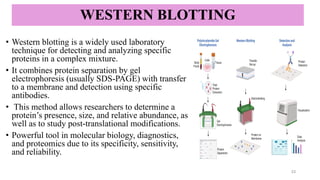
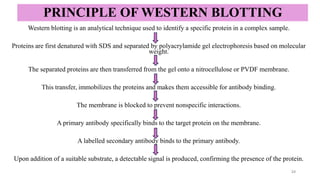
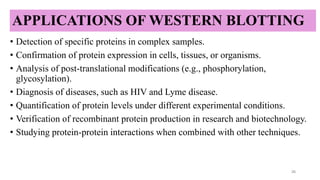
Western blotting-a specific technique used to detect and identify target
proteins using antibody–antigen interactions.












![REFERENCE
• Kurien & Scofield; Scofield, RH (2006). "Western Blotting". Methods 38 (4): 283–293. Burnette WN. (1981). "'Western
blotting': electrophoretic transfer of proteins from sodium dodecyl sulfate—polyacrylamide gels to unmodified
nitrocellulose and radiographic detection with antibody and radio iodinated protein A". Analytical Biochemistry 112 (2):
195 203.
• Fajriyah R. Paper review: An overview on microarray technologies. Bulletin of Applied Mathematics and Mathematics
Education. 2021;1(1):21–30. doi:10.12928/bamme.v1i1.3854.
• Kumar A, Goel G, Fahrenbach E, Puniya AK, Singh K. Microarrays: the technology, analysis and application.
Engineering in Life Sciences. 2006;6(5):451–459.
• Kurien BT, Scofield RH. Western blotting: an introduction. In: Kurien BT, Scofield RH, editors. Western blotting:
methods and protocols. New York (NY): Humana Press; 2015. p. 17–30.
• Aydin S, Emre E, Ugur K, Ata M, Aydin I, Sahin I, et al. An overview of ELISA: A review and update on best laboratory
practices for quantifying peptides and proteins in biological fluids. Journal of International Medical Research.
2025;53(2):1–18. doi:10.1177/03000605251315193.
• Idowu OS, Loko DO, Ogundipe SO, Mensah E. Optimizing SDS-PAGE for accurate protein characterization in
nutritional research and food quality assessment. Int J Innov Sci Res Technol. 2025;10(1).
doi:10.5281/zenodo.14744563.
• Sedighi A, Li PCH. Challenges and future trends in DNA microarray analysis. In: Li PCH, editor. Microarray
technology: methods and applications. New York (NY): Springer; 2010. p. 25–44.
• ELISA-Antibody.com. ELISA introduction: ELISA principle [Internet]. ELISA-Antibody; [cited 2026 Jan 31]. Available
from: http://elisa-antibody.com/ELISA-Introduction/ELISA-Principle.html
• Vayanaperumal K. Western blotting: a review: principle, protocol and problem solving. GSC Advanced Research and
Reviews. 2023;16(3):178–187. doi:10.30574/gscarr.2023.16.3.0324
29](https://image.slidesharecdn.com/cmp-260210154429-659b9890/85/ELISA-MICRO-ARRAY-TECHNIQUE-SDS-PAGE-WESTERN-BLOTIING-29-320.jpg)
