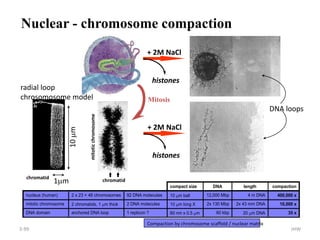
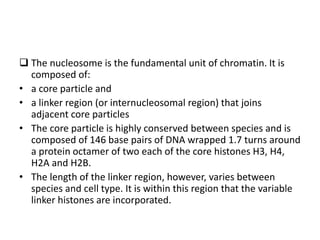
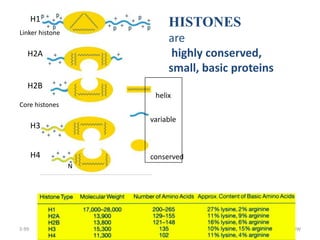
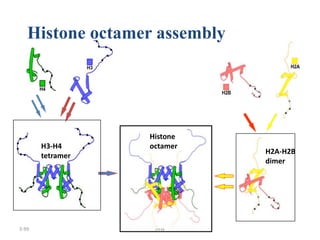
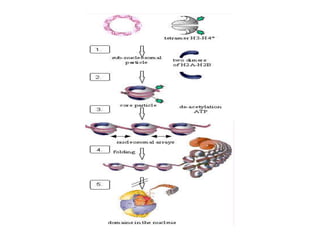
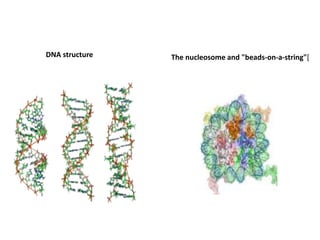
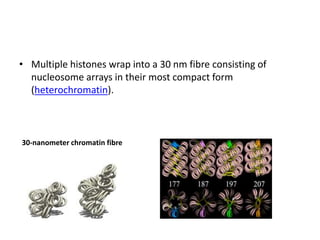
The document discusses micromolecules, specifically focusing on chromatin, its structure, and functions. It explains the roles of nucleosomes and histones in packaging DNA within the cell nucleus, along with the hierarchical organization of chromatin. The conclusions highlight the significance of chromatin packaging in regulating genome function and the ongoing exploration of post-translational histone modifications.