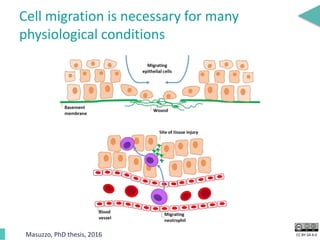
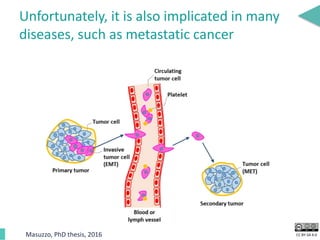
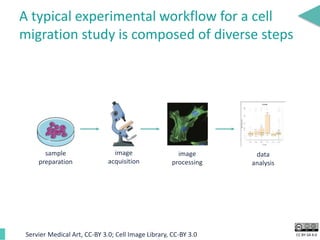
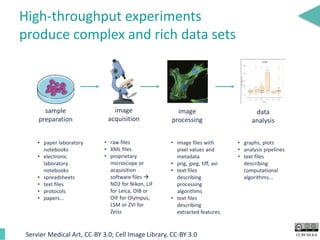
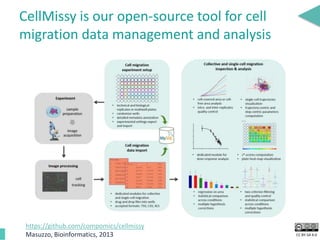
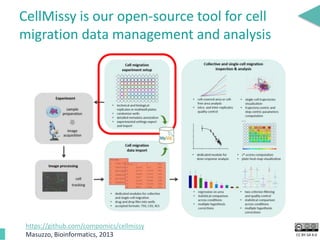
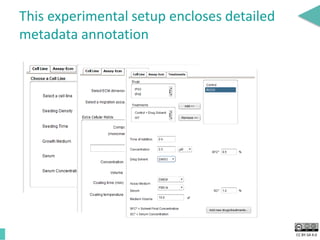
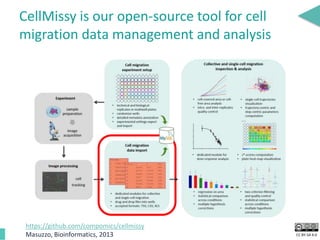
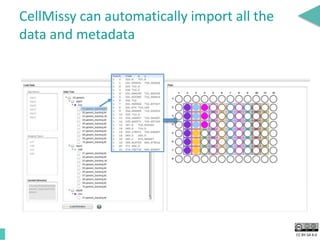
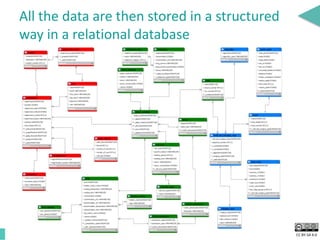
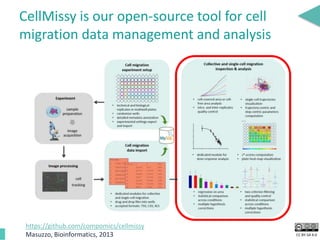
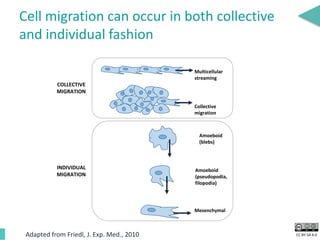
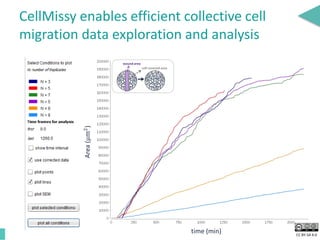
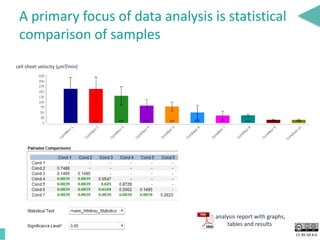
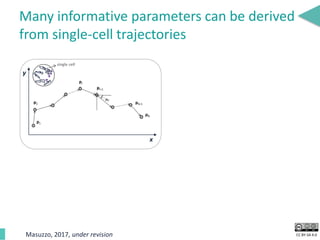
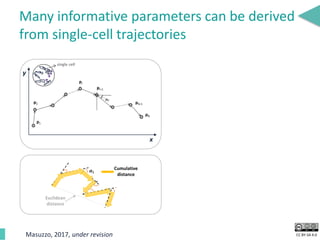
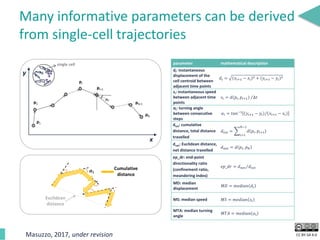
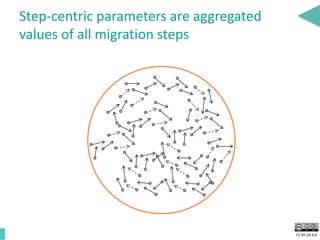
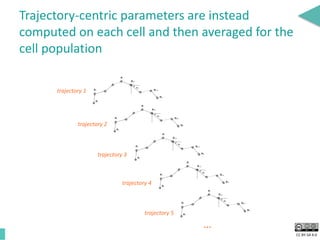
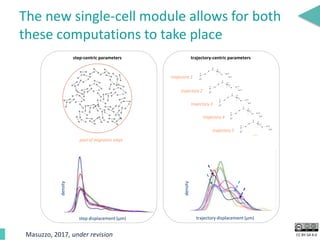
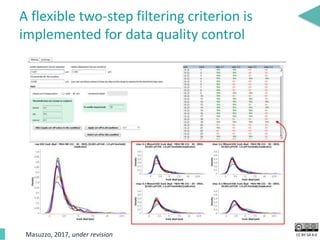
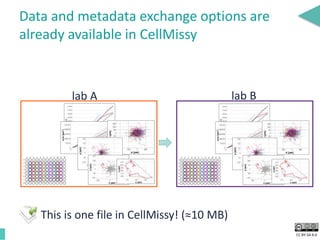
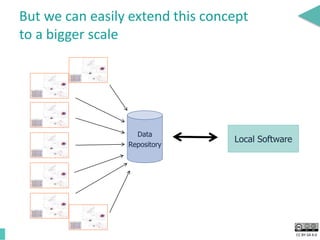
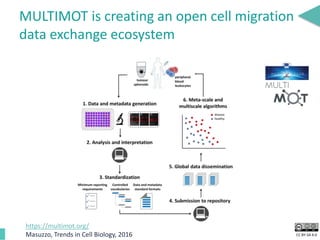
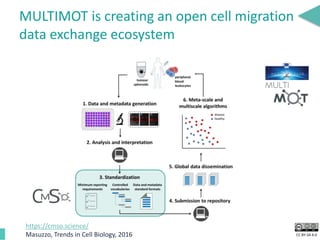
This document discusses the development of an open data exchange ecosystem for cell migration research. It describes CellMissy, an open-source software tool for managing and analyzing cell migration data. CellMissy allows researchers to capture experimental metadata, import diverse data types into a structured database, and perform both collective and single-cell analysis. The document proposes extending CellMissy's data sharing capabilities into a larger repository called MULTIMOT to facilitate global exchange of cell migration data as part of the broader open science movement.