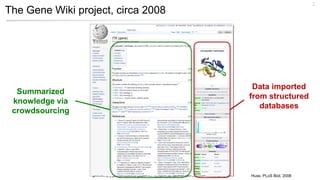
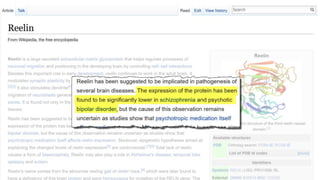
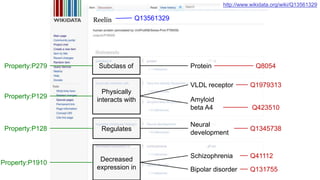
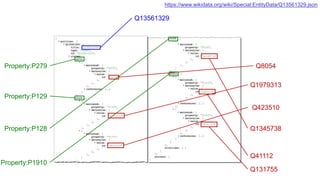
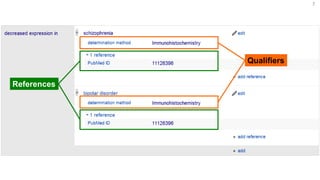
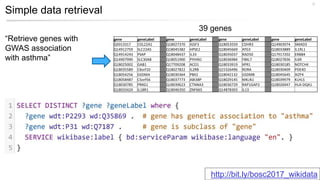
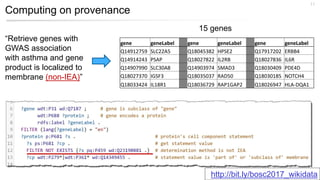
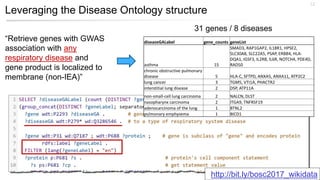
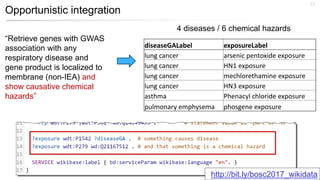
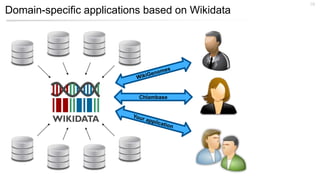
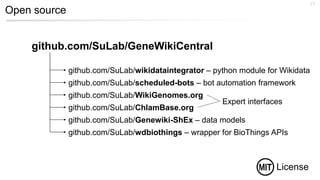
This document discusses using Wikidata as an open, community-maintained database of biomedical knowledge. It describes how Wikidata allows data to be imported from structured databases and knowledge to be summarized via crowdsourcing. Examples are given of simple data retrieval queries around genes associated with asthma and integrating additional constraints. The potential for domain-specific applications is discussed, with Chlambase.org given as an example for the Chlamydia research community. Open source tools and opportunities for the community to contribute are also mentioned.