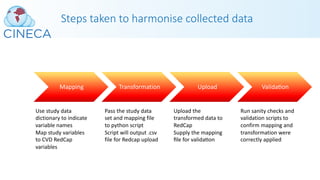
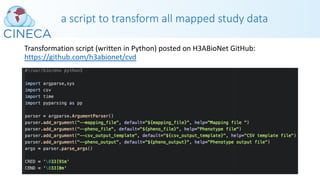
This webinar discussed H3ABioNet's experiences with phenotype data harmonization and standards development. It covered both retrospective and prospective harmonization efforts for the H3Africa consortium. Retrospective harmonization involved mapping variables from six cardiovascular disease studies to develop a harmonized CHAIR resource, while prospective efforts involved developing a standardized core set of phenotypes and modules for new studies. Many challenges were encountered, but lessons were learned regarding communicating standards, obtaining clean data, and respecting participant populations. The webinar provided an overview of the harmonization process and acknowledged contributions to the work.


![Today’s presenter
Katherine Johnston is currently co-chair of the H3ABioNet Health InformaPcs
Work Package and H3Africa Phenotype HarmonisaPon Working Group, focused
on data harmonisaPon efforts and building Phenotype Standards in the African
context. She provides support to the H3Africa ConsorPum studies in phenotype
data collecPon, harmonisaPon, soware pla]orms and incorporaPon of
ontologies. With a long-standing desire to play a role in advances in genePc
research, Katherine moved from a posiPon as Head Clinical Data Manager at
AHRI, Durban and joined the ComputaPonal Biology Division at UCT, Cape Town
as an H3ABioNet Soware Developer. She completed an Honours degree in
Computer Science and Economics and started her career working in Electronic
Health Record systems development.
Discovering a passion for helping medical professionals in the field of patient care by building them useful
electronic tools she joined CAPRISA as a Clinical Data Manager in 2004 and worked with enjoyment in the
field of clinical data management for 13 years. During this time, she married and had three beautiful
children, the first of whom has Down Syndrome. As a result, she finds herself unexpectedly combining her
interest in genetics, health informatics and patient care with her love for her children, advocating for
inclusivity and research advancements in medical care, data collection standards and software development
to benefit all genetically diverse people in Africa.](https://image.slidesharecdn.com/cinecah3abionetwebinarslides-200204103734/85/CINECA-webinar-slides-H3ABioNet-Experiences-in-Phenotype-Data-Harmonisation-and-Standards-Development-5-320.jpg)












![Many challenges were met…
With the retrospecPve data harmonisaPon:
• Did parPcipant consent forms cover meta-analysis? What DUO codes applied?
• Could fragmented IRBs and insPtuPons in different countries agree on harmonised efforts?
• Where would curated and harmonised data reside?
• IRB approval for harmonisaPon database had to be obtained.
• Diverse or lacking data governance policies in different African countries created uncertainty.
• Tribal poliPcs and minority populaPons had concerns about data sharing and access that minimised data that was
allowed to be harmonised.
• Who could send or ship samples out a country – was it permiked?
• Different data pla]orms and hetrogeneity of phenotype data meant each study had to be manually mapped to
harmonised variables.
• Studies were slow to completely clean and analyse data ready for meta-analysis
• Visibility of some data were a concern e.g. site – some minority populaPons were concerned about idenPficaPon and
potenPal risks.
• Determining whether or not to include longitudinal data and how
• RetrospecPve coding of ethnicity/tribe and language needed to be done
• Calculated variables had to be done at a study level – the generic script could not accommodate different scenarios from
different studies to where mulPple fields informed a single harmonised data element.](https://image.slidesharecdn.com/cinecah3abionetwebinarslides-200204103734/85/CINECA-webinar-slides-H3ABioNet-Experiences-in-Phenotype-Data-Harmonisation-and-Standards-Development-26-320.jpg)





