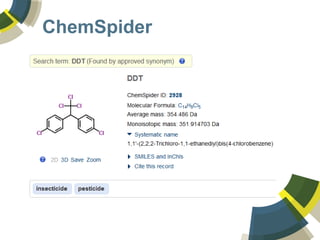
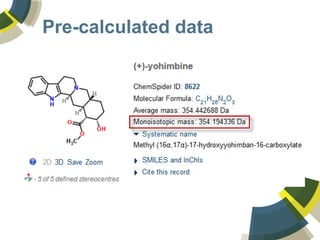
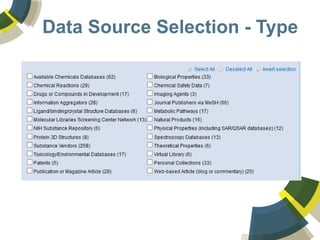
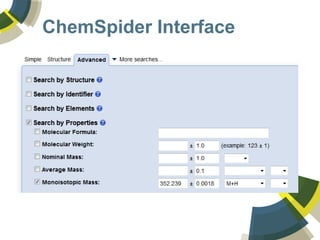
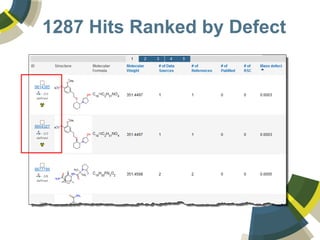
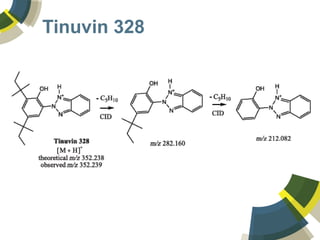
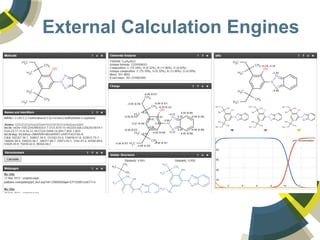
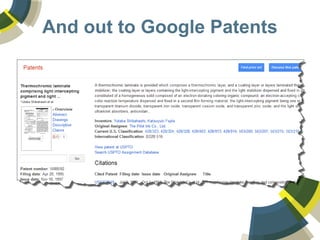
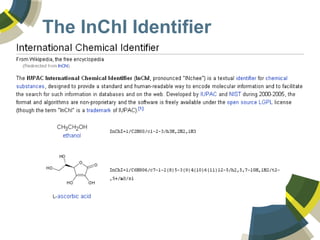
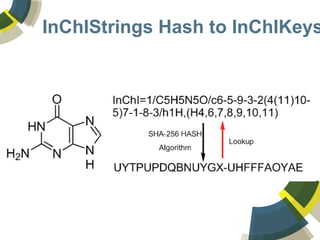
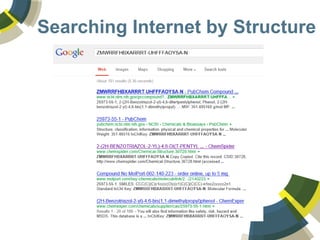
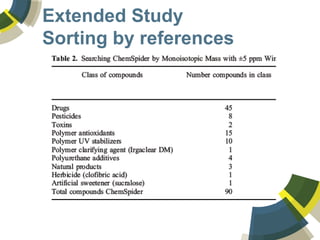
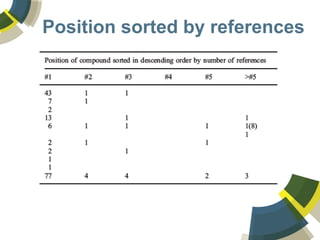
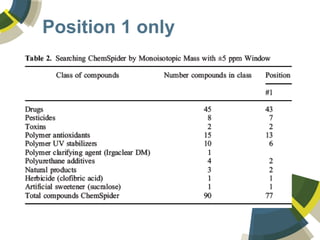
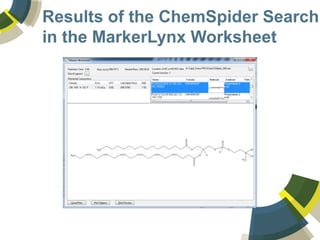
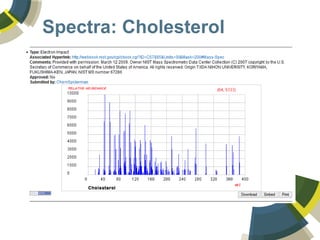
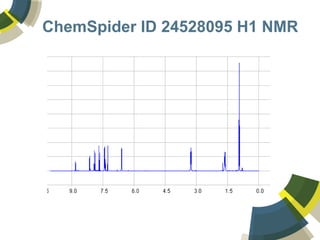
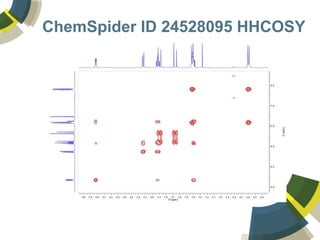
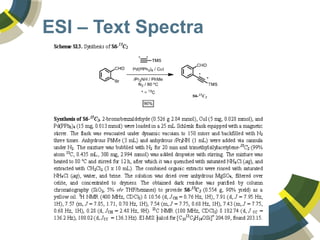
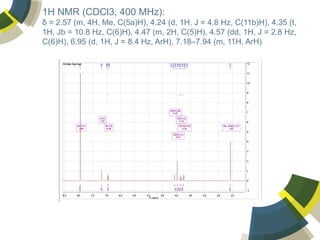
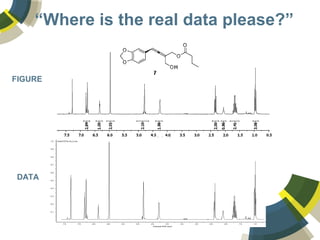
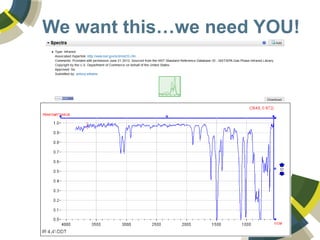
The document discusses the use of ChemSpider, an online chemistry database, for structure identification in mass spectral data, providing free access to over 34 million chemicals. It highlights the database's capabilities in searching by mass, linking structure-based data, and offering programming interfaces for integration. Future developments aim to expand spectral data availability, including the transformation of text and figures into spectral formats.