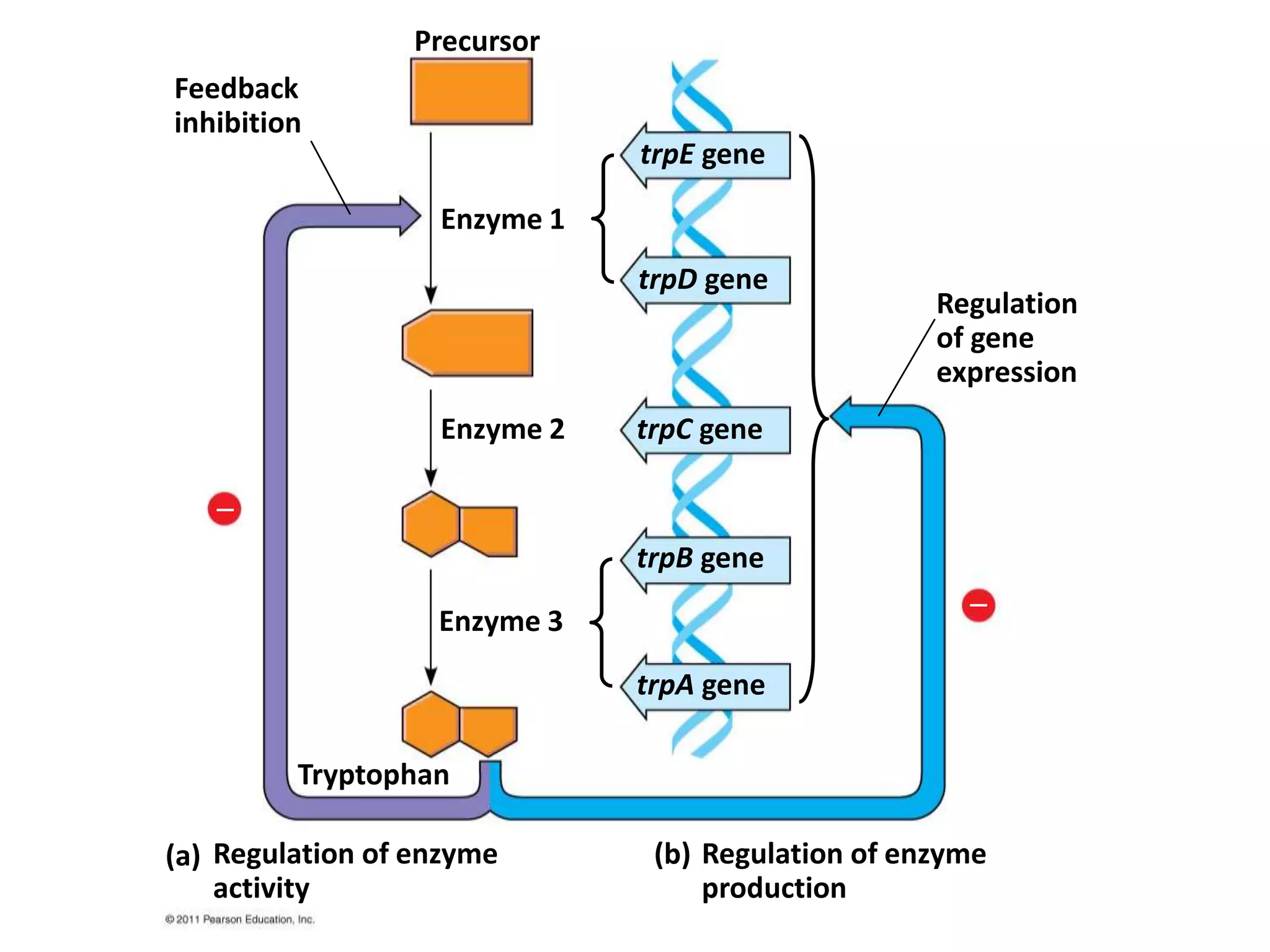
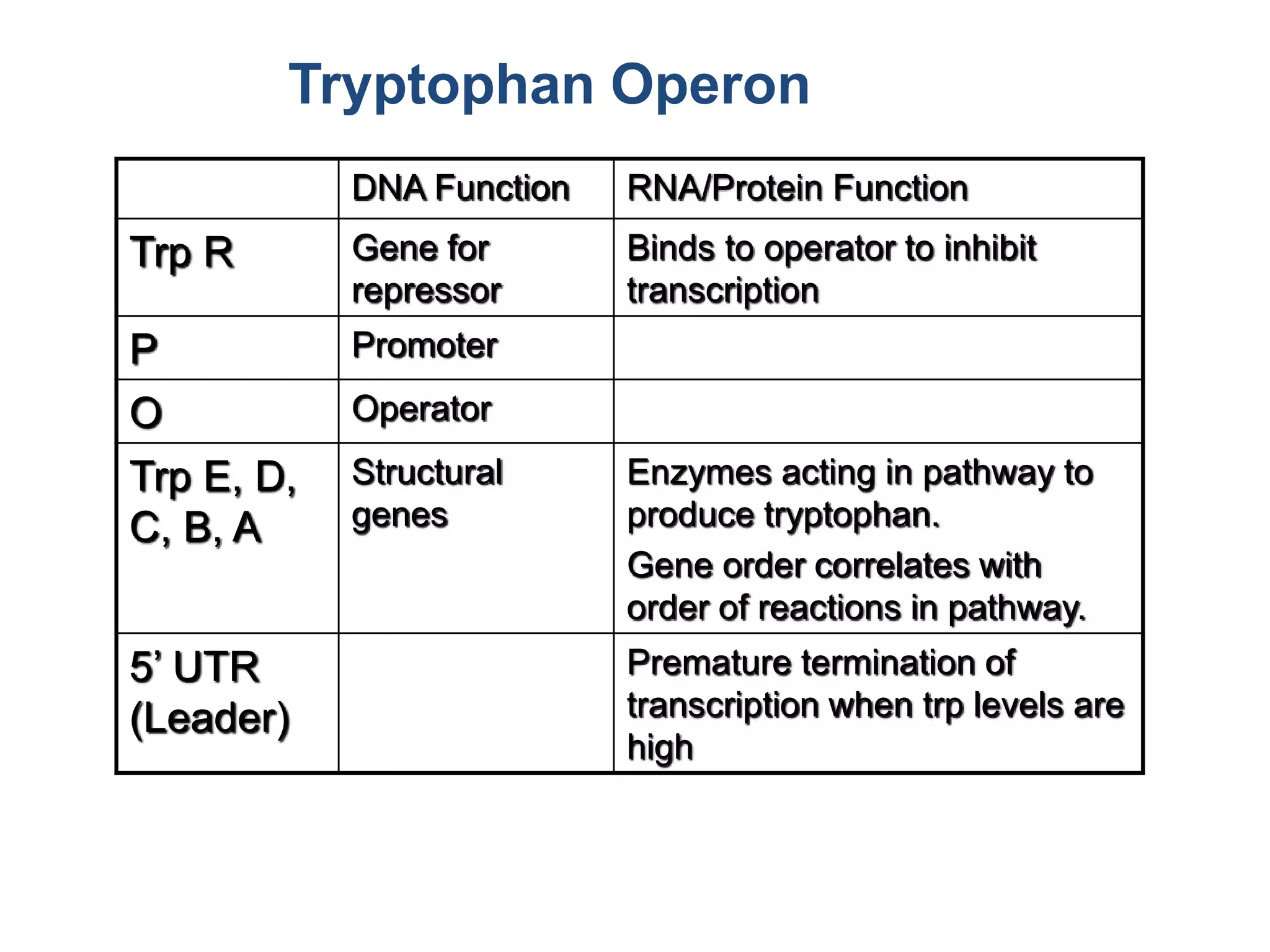
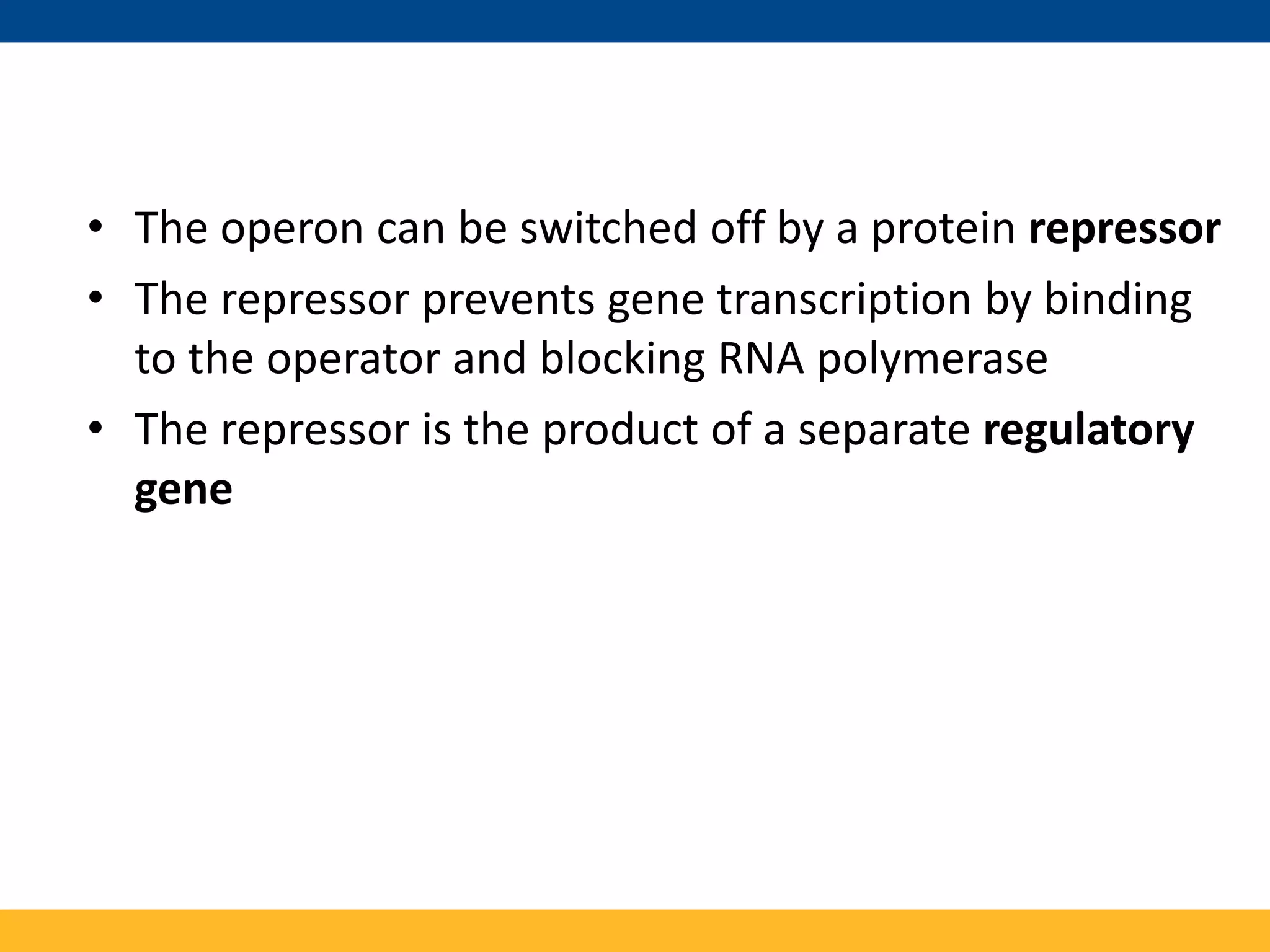
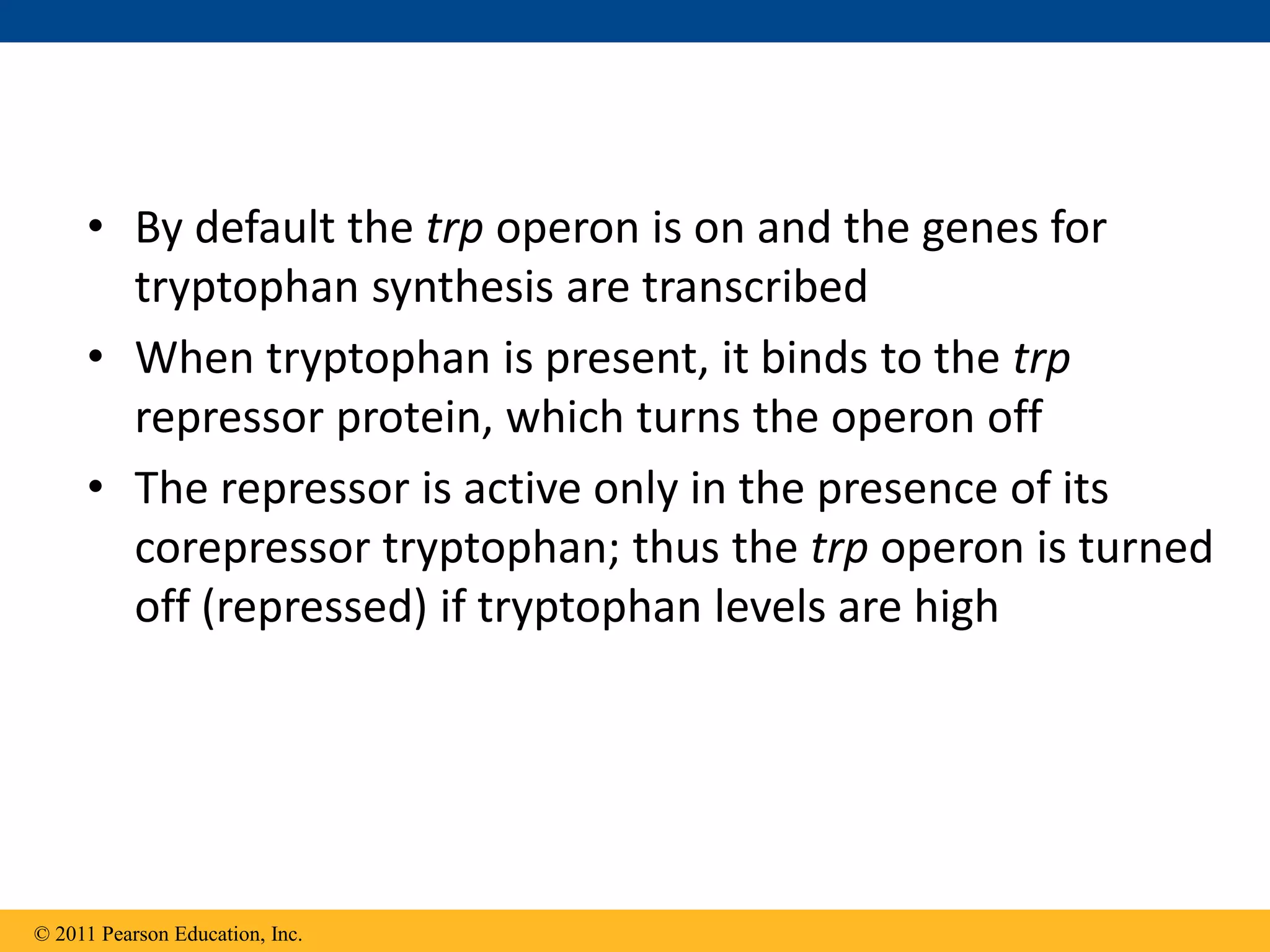
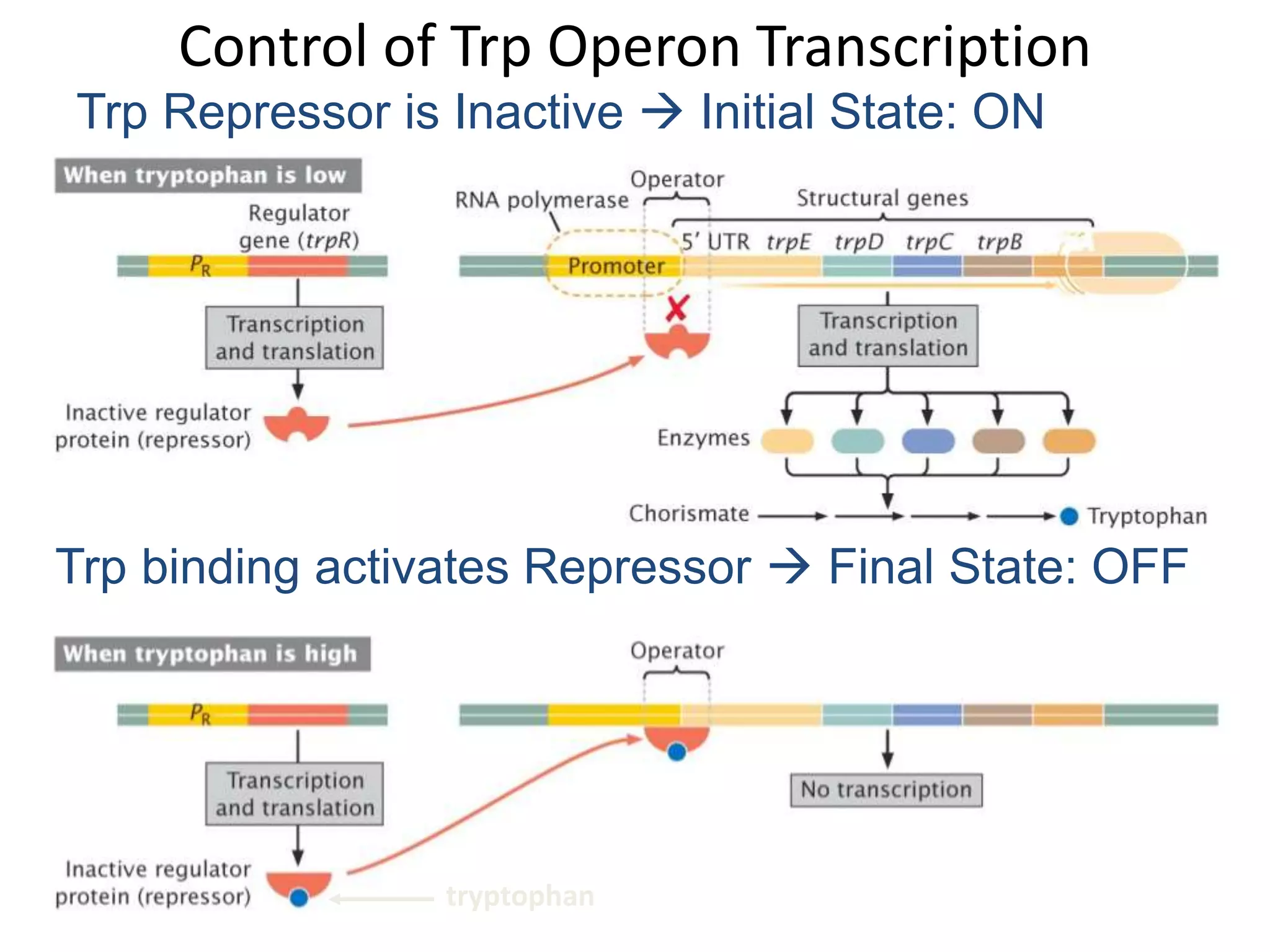
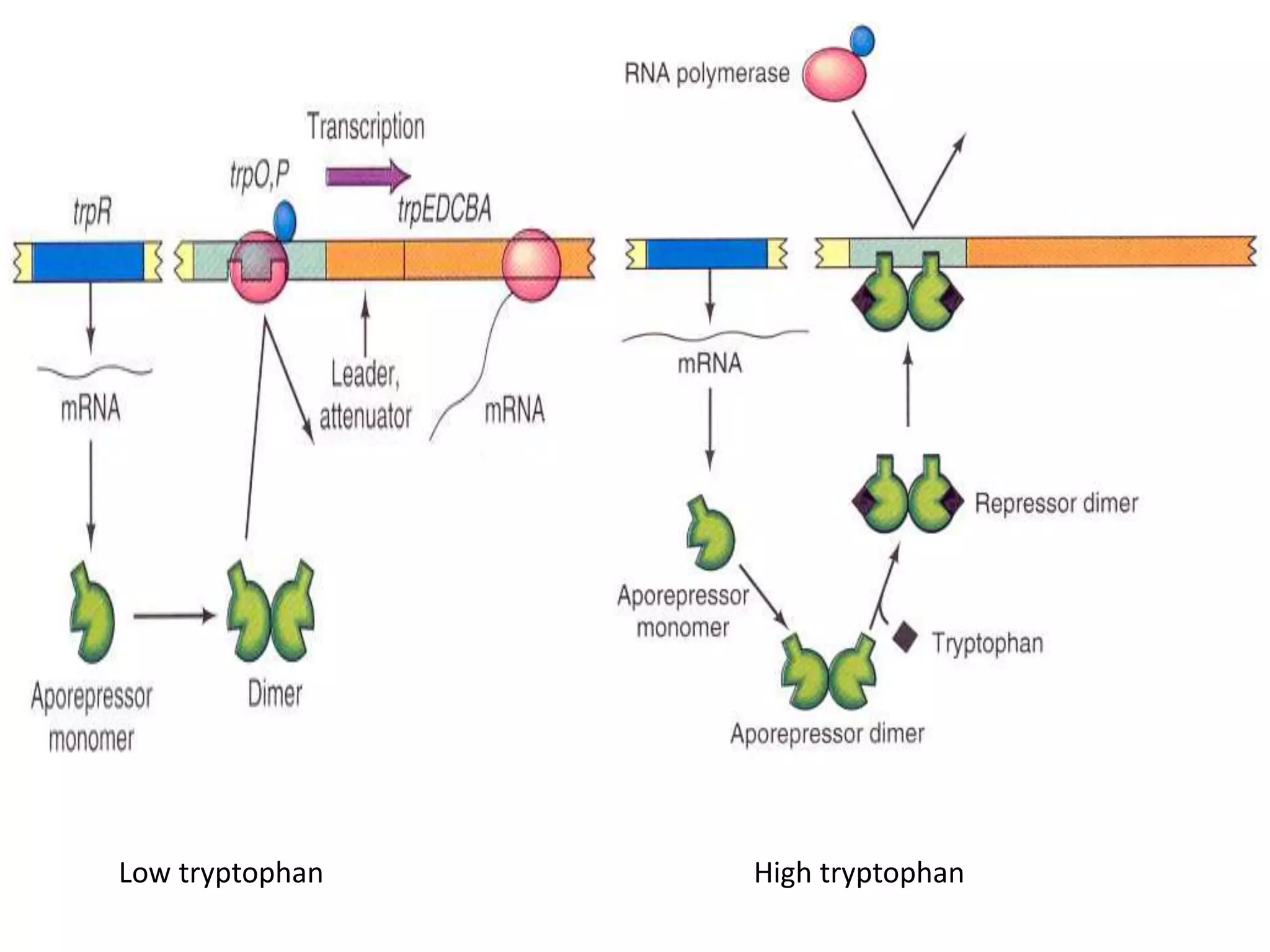
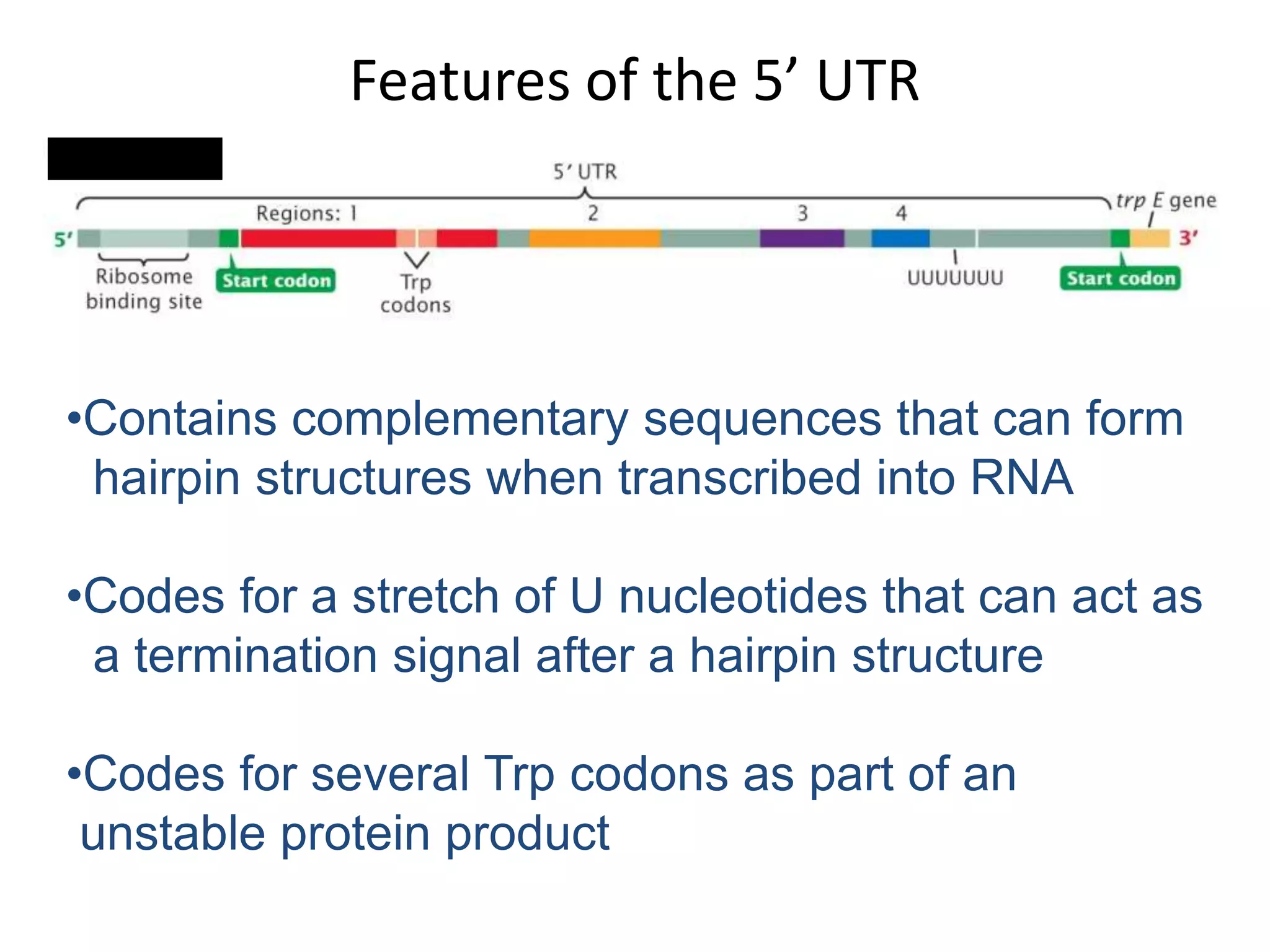
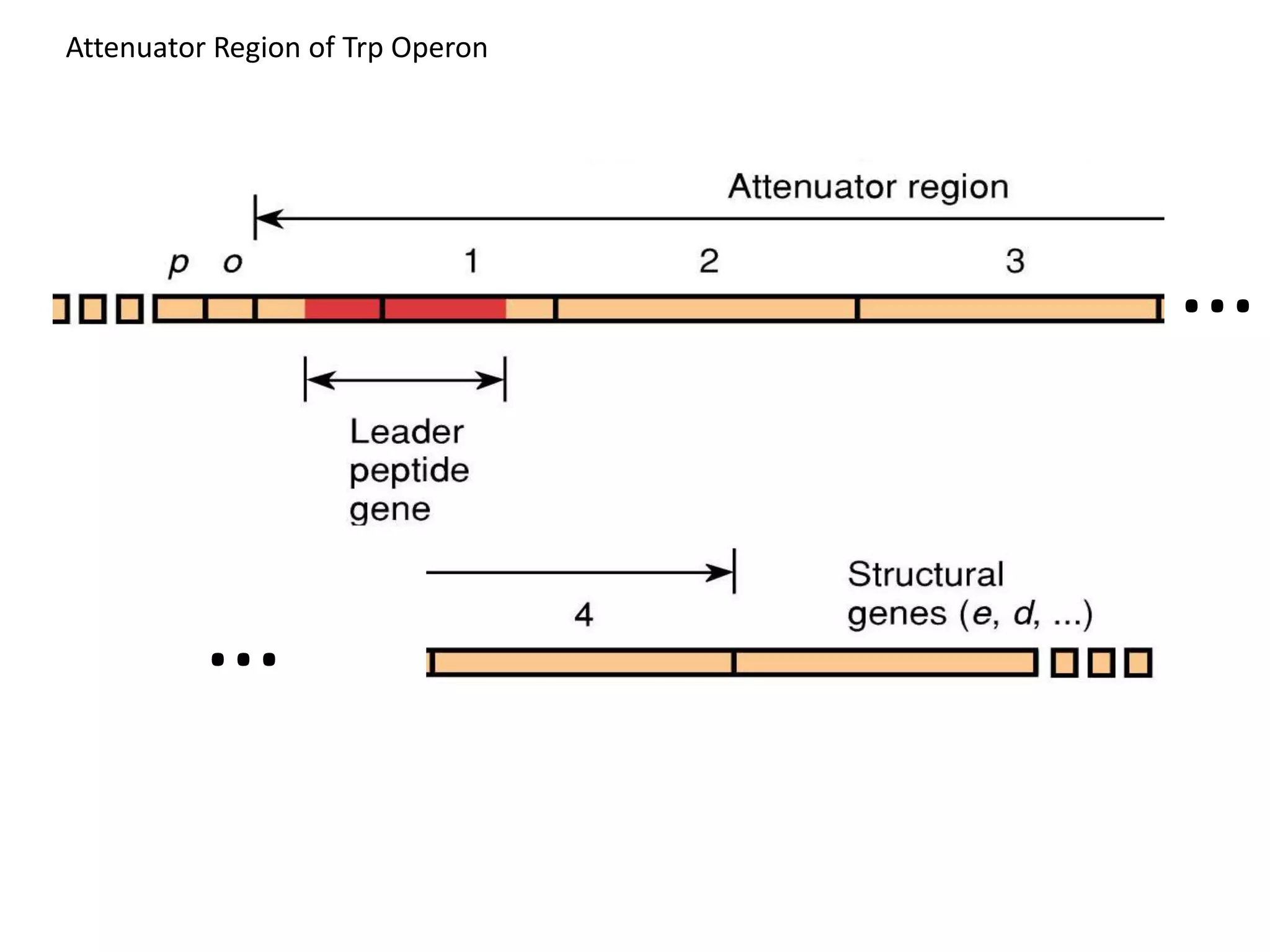
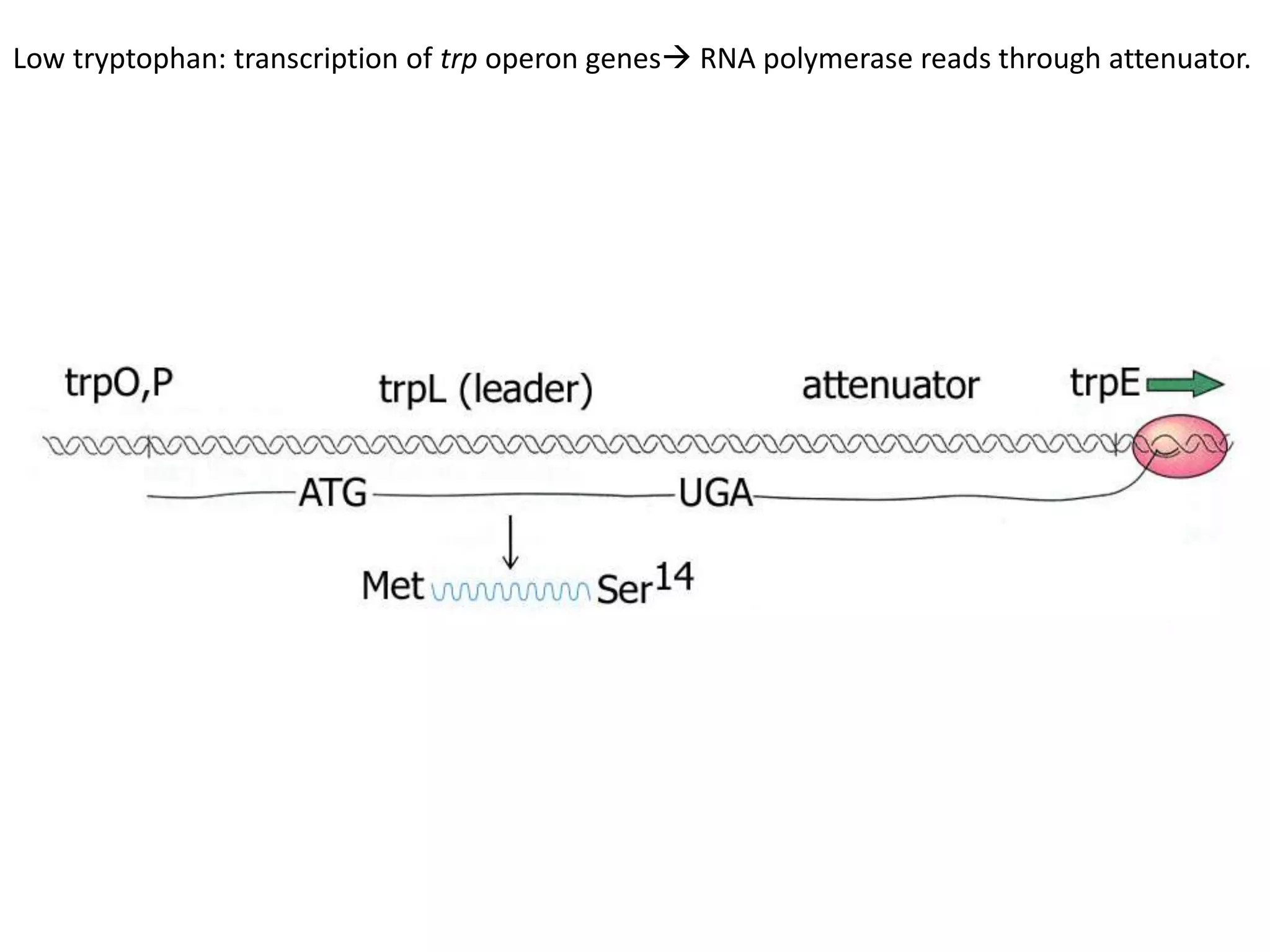
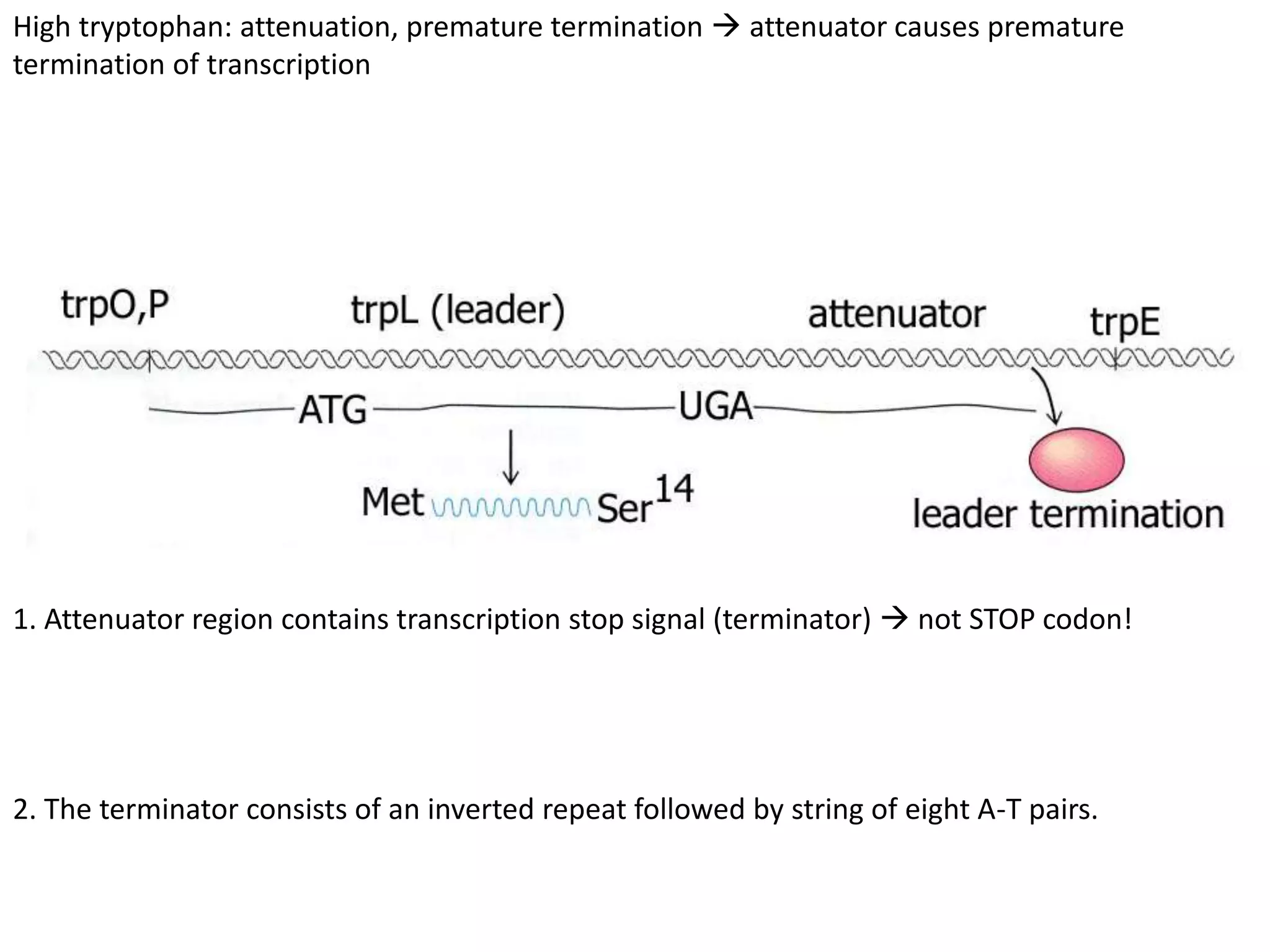
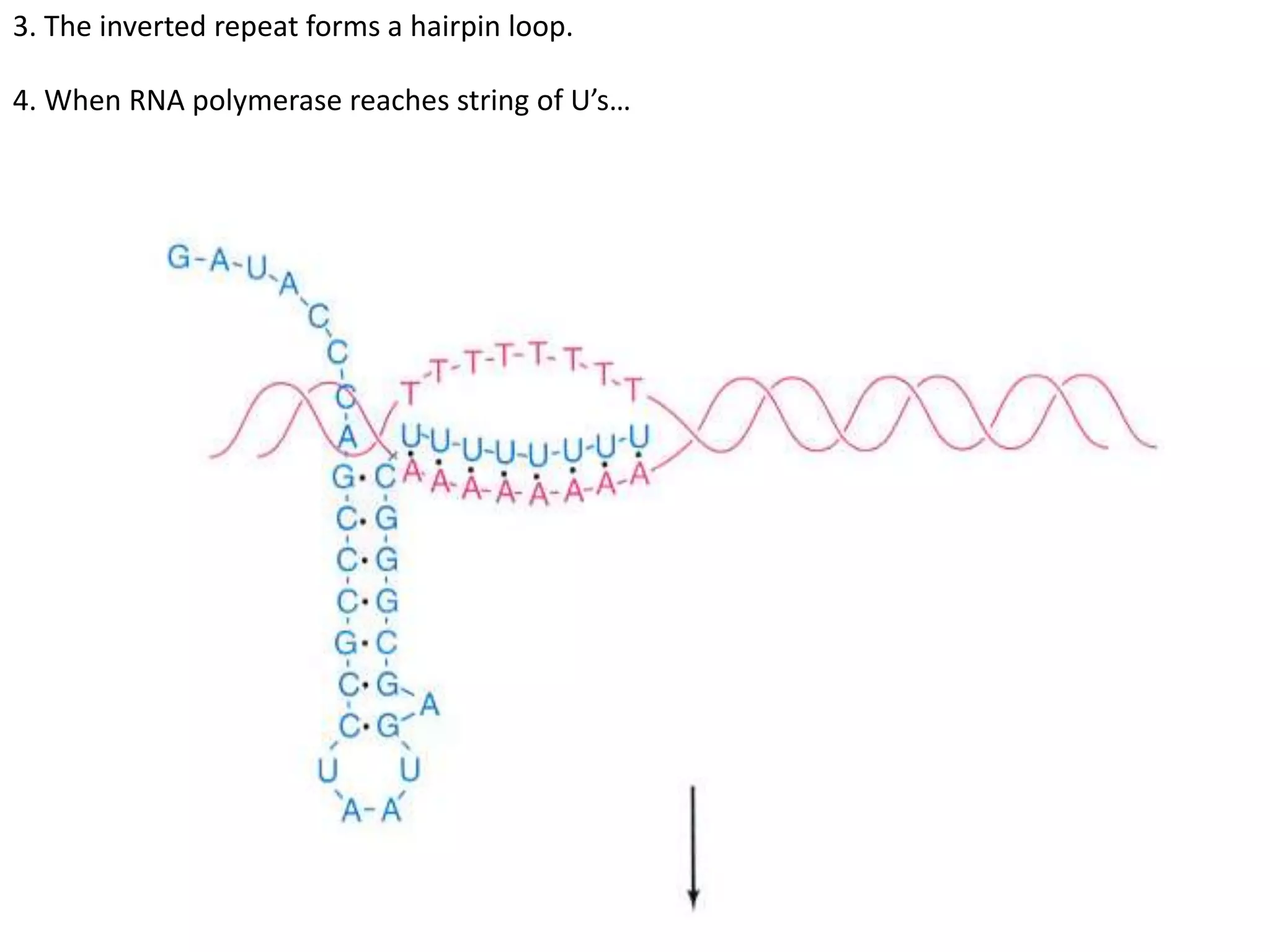
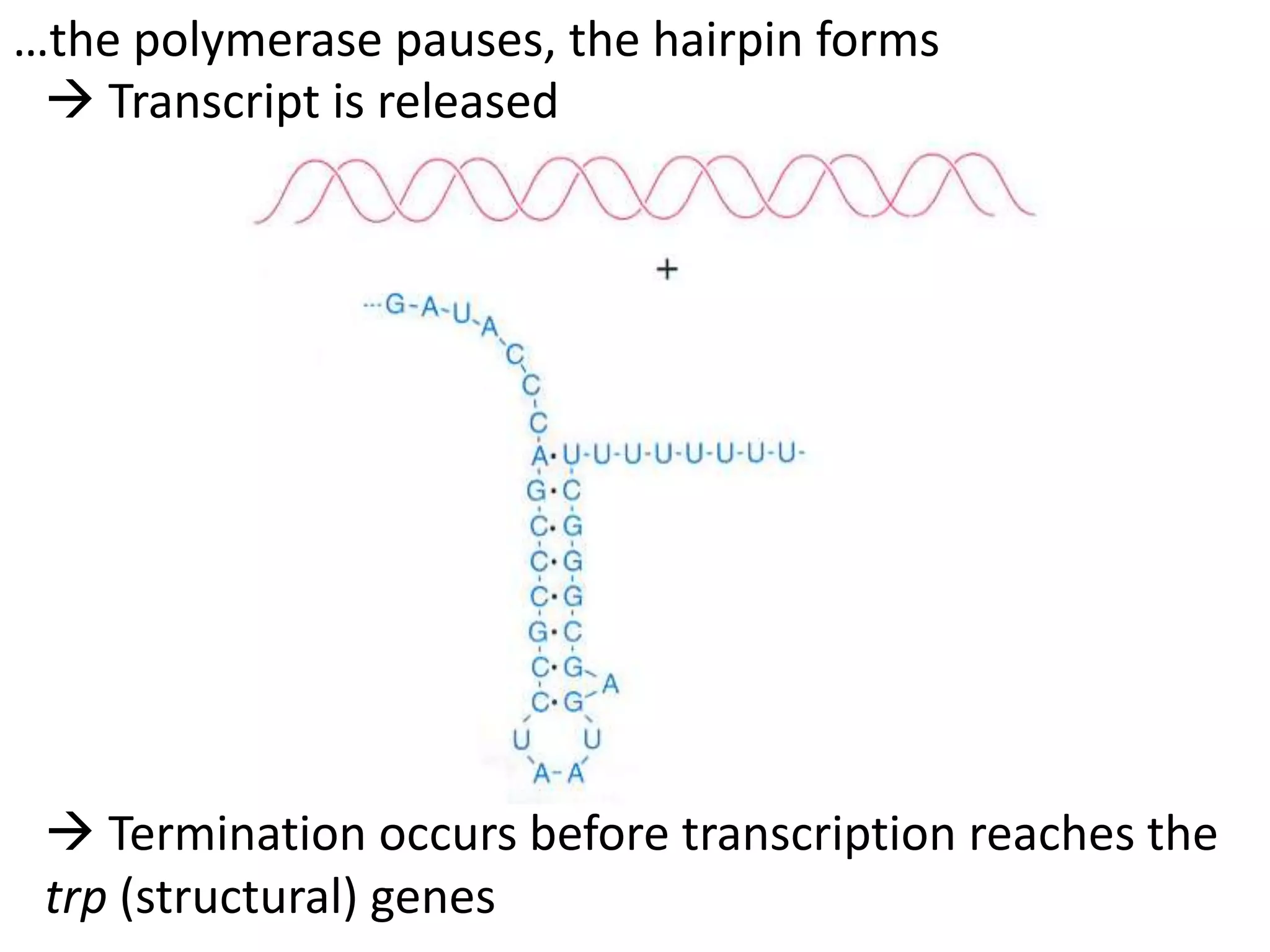
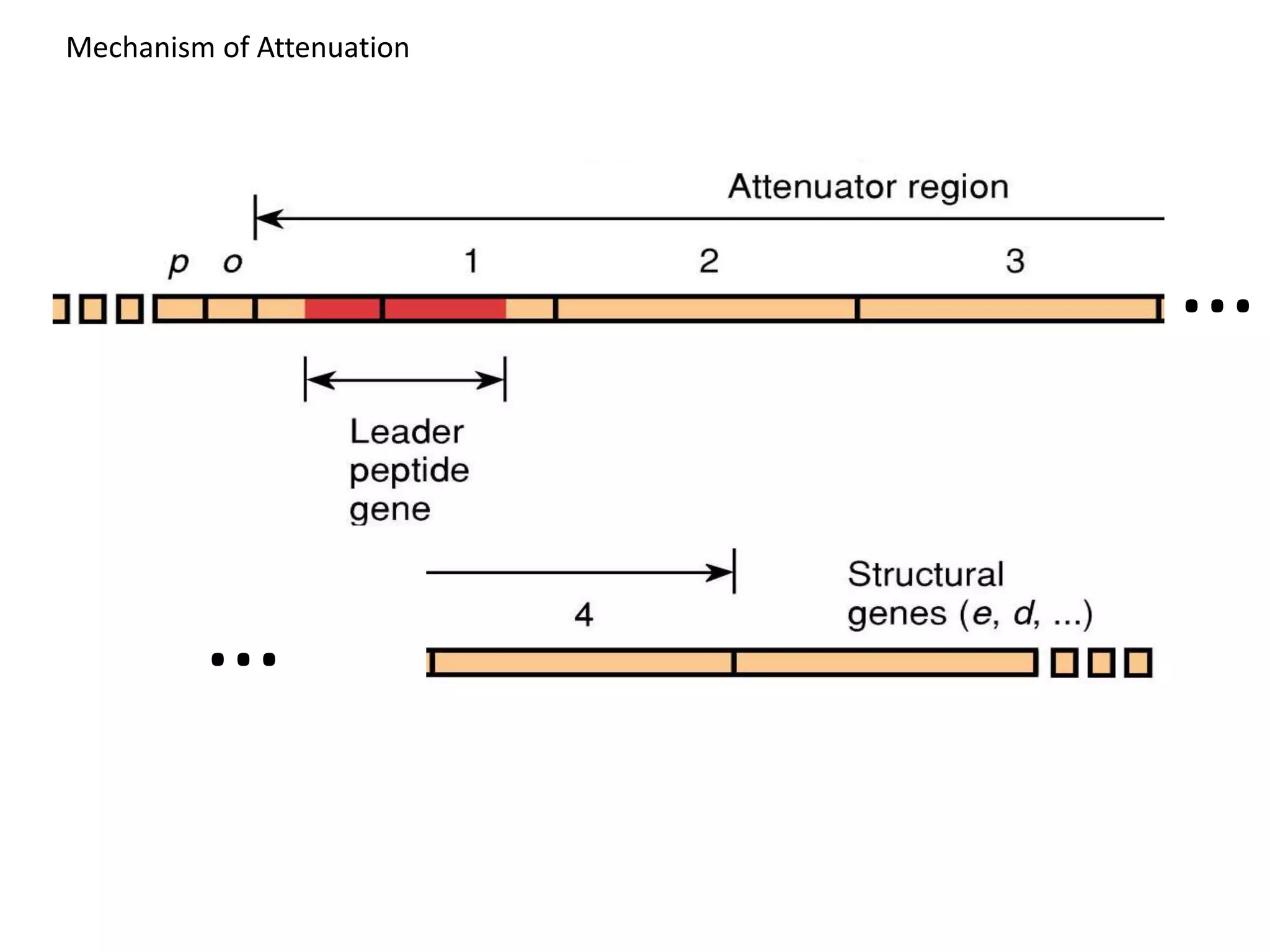
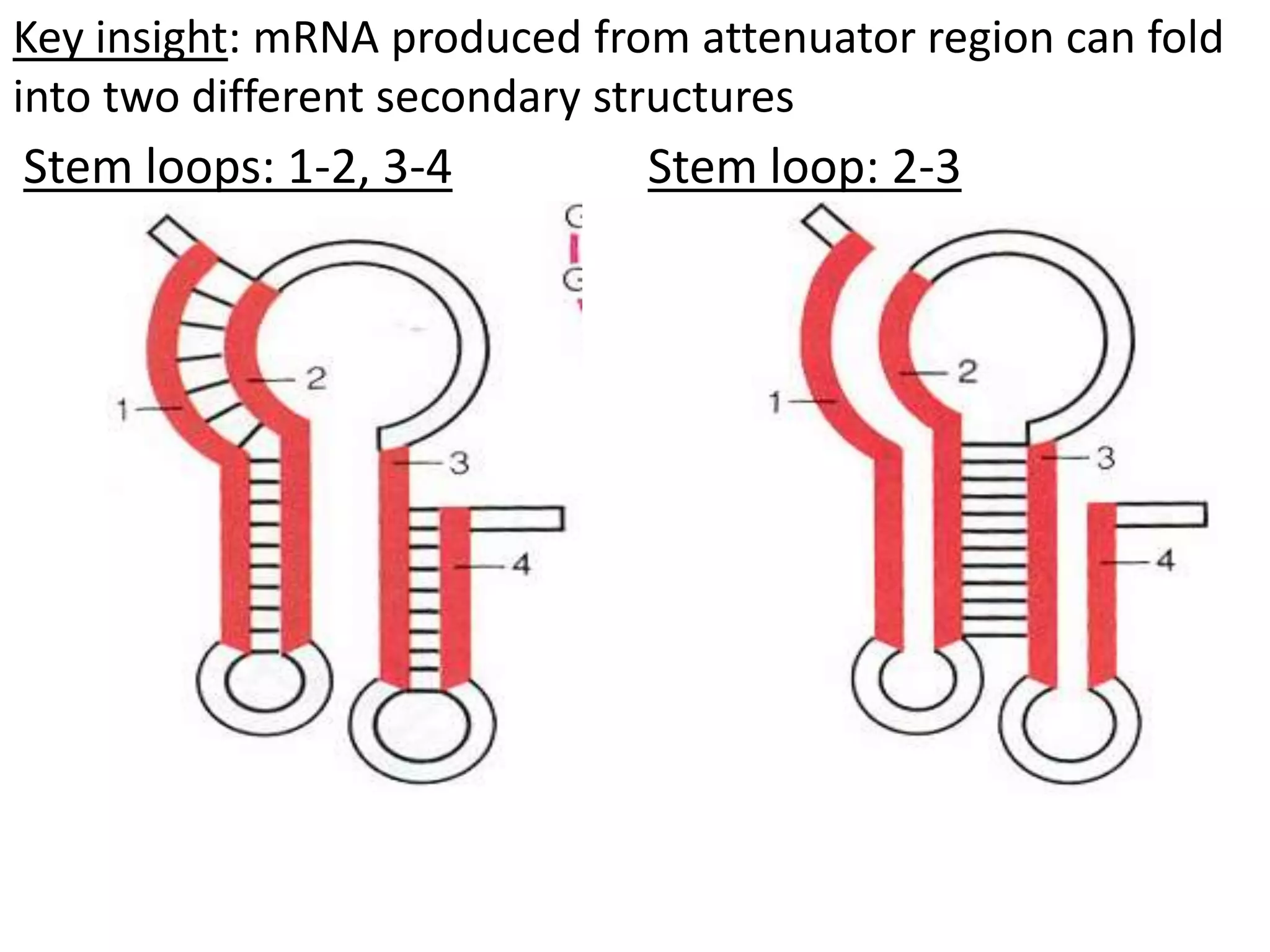
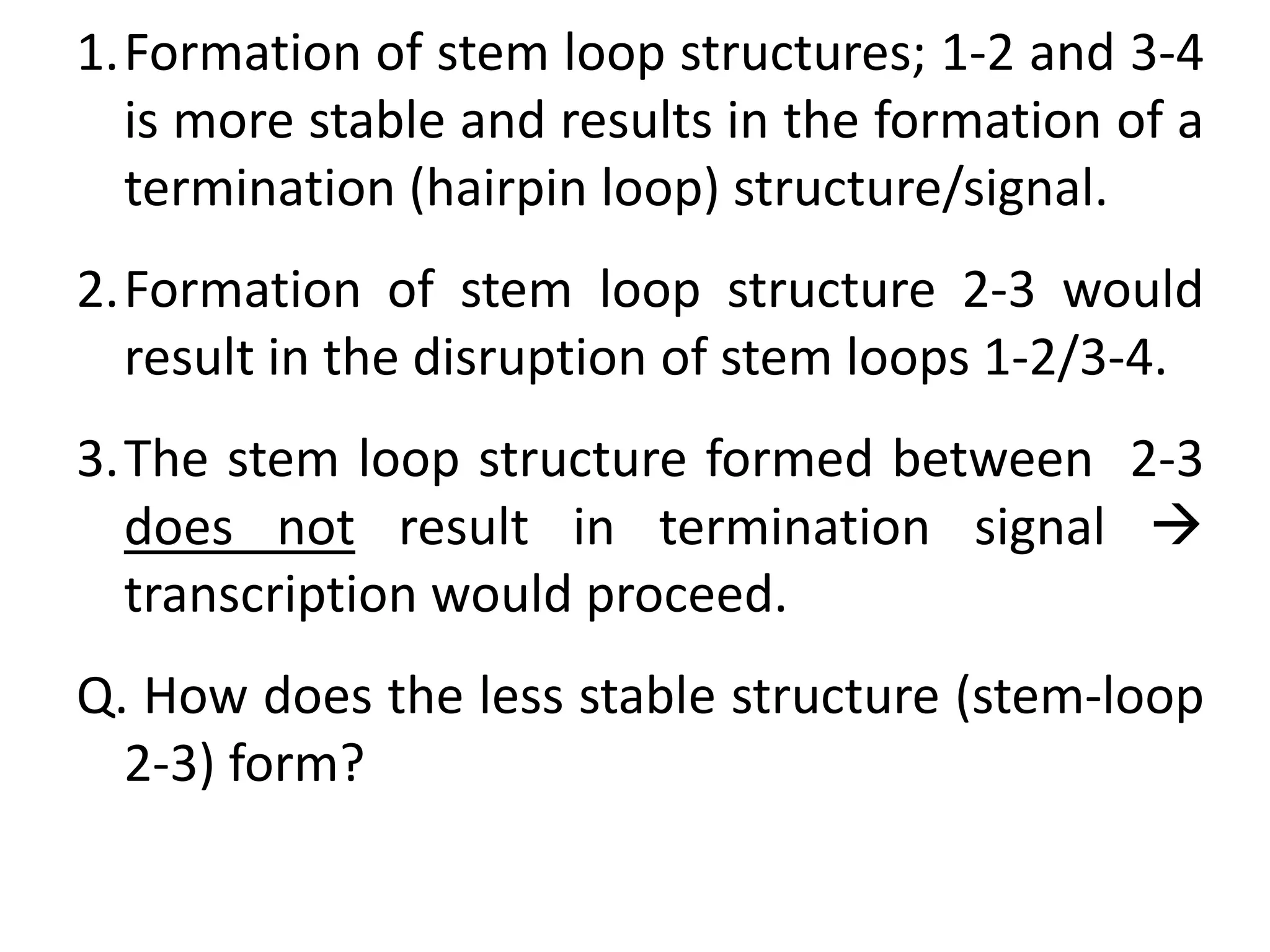
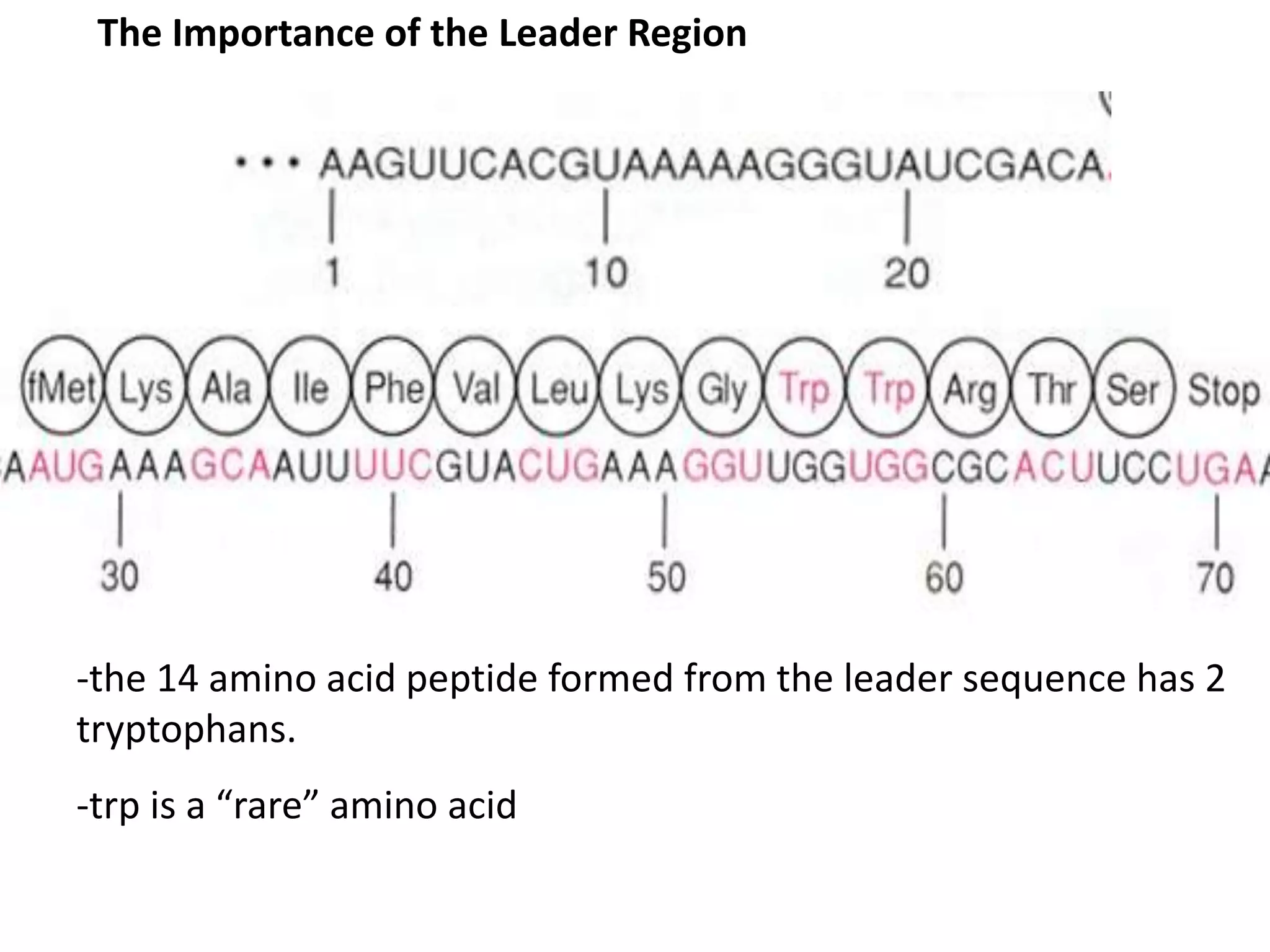
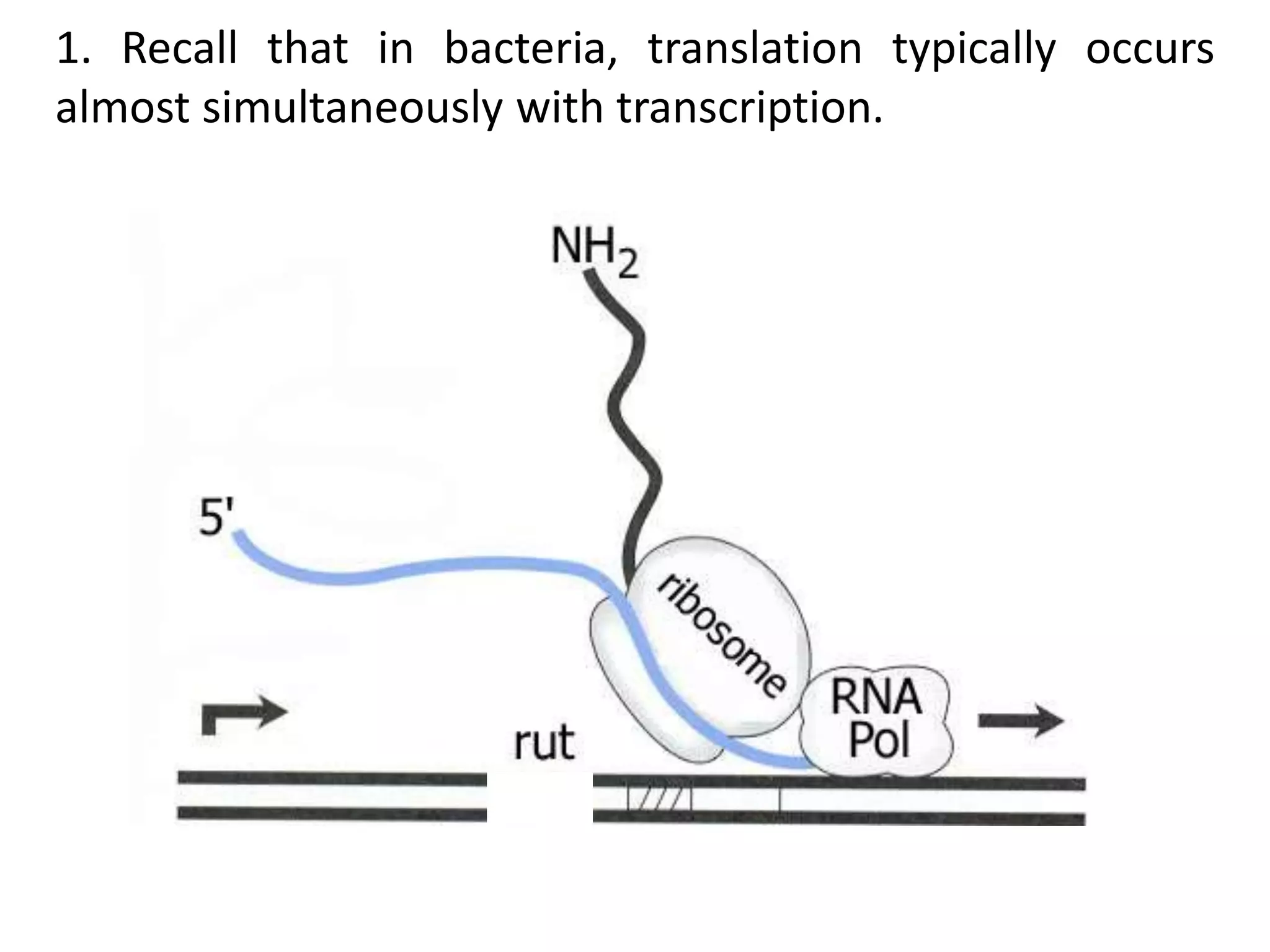
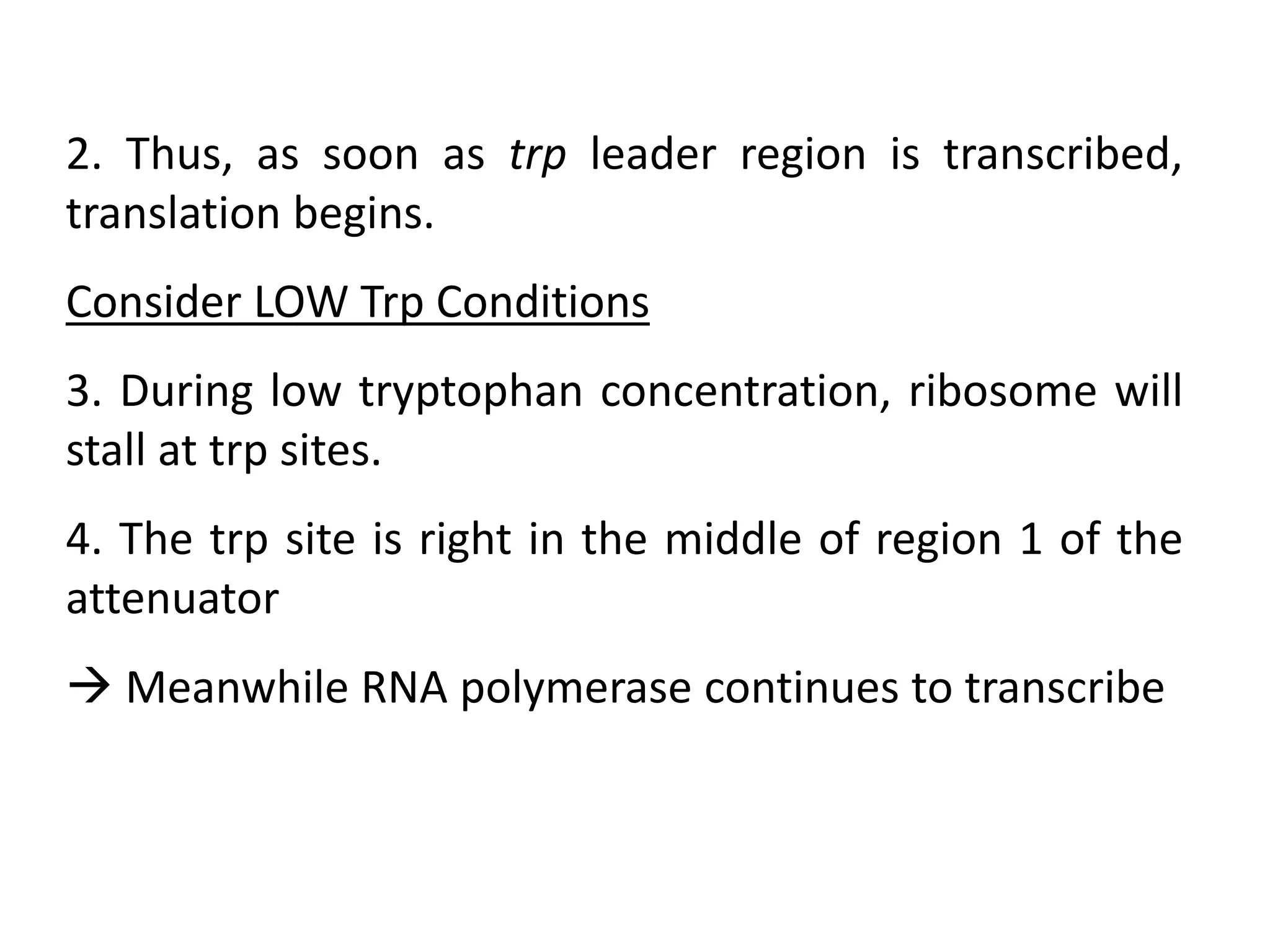
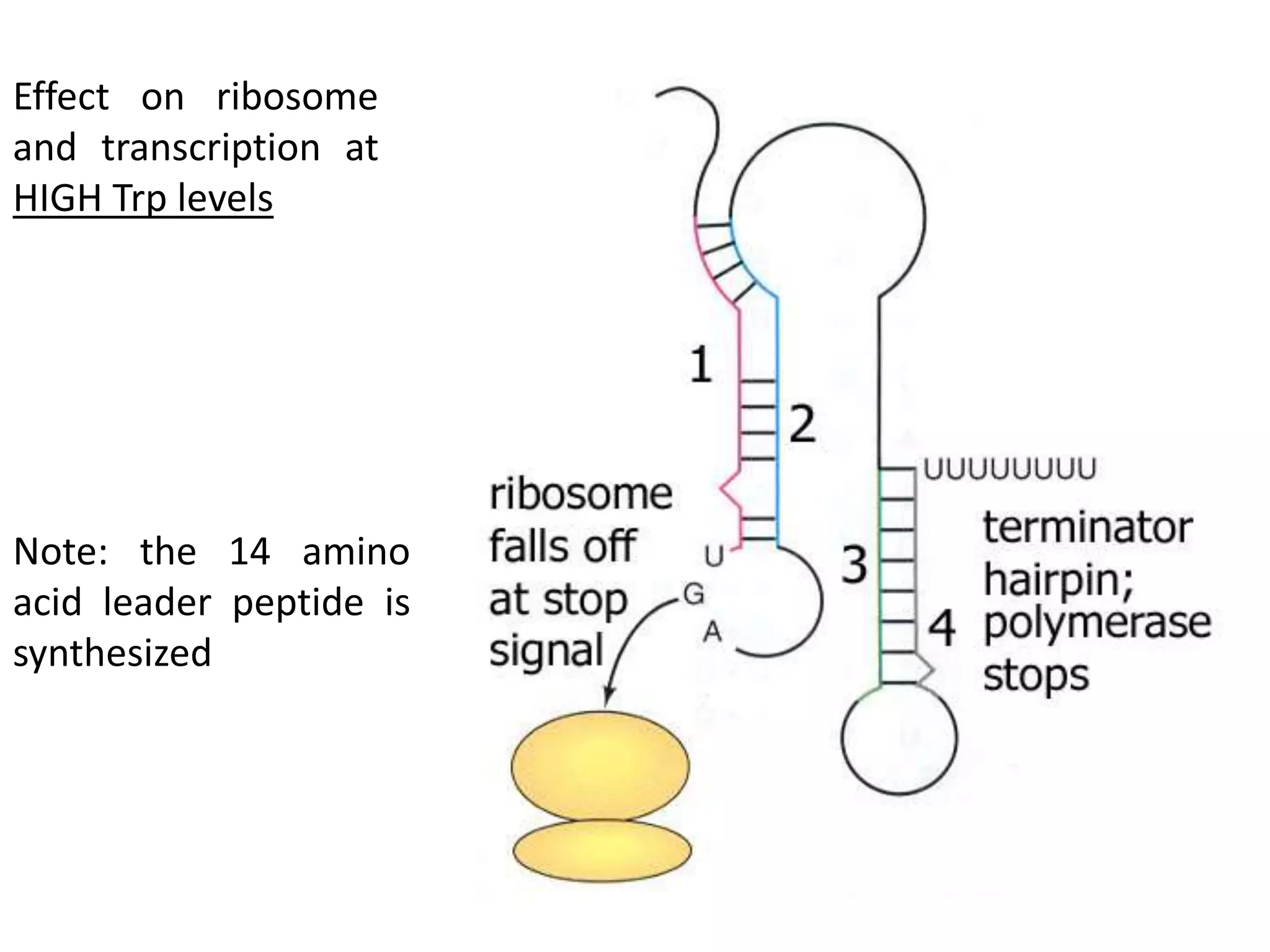
The tryptophan (trp) operon regulates gene expression in response to intracellular tryptophan levels through both repression and attenuation. When tryptophan is low, the trp repressor is inactive and transcription proceeds unimpeded. Additionally, ribosome stalling at trp codons in the 5' UTR prevents transcriptional termination through attenuation. Together, this maximizes production of tryptophan synthesis enzymes. When tryptophan is high, it activates the trp repressor which binds DNA and inhibits transcription. It also enables fast ribosome transit through the 5' UTR, allowing an alternative RNA structure that signals premature termination before the enzyme genes.