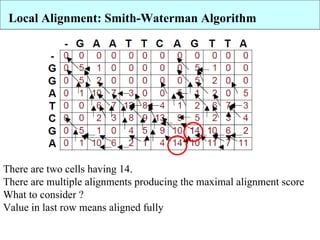
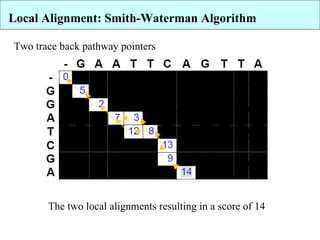
The document discusses algorithms for optimal sequence alignment using dynamic programming. It describes the Needleman-Wunsch algorithm for global alignment and the Smith-Waterman algorithm for local alignment. Both algorithms use dynamic programming to find the highest scoring alignment between two sequences by dividing the problem into independent subproblems. The Needleman-Wunsch algorithm allows gaps across the entire length of the sequences, while Smith-Waterman only considers local regions with high similarity.
![S.Prasanth Kumar, Bioinformatician Genomics Sequence Alignment : Complete Coverage-I S.Prasanth Kumar Dept. of Bioinformatics Applied Botany Centre (ABC) Gujarat University, Ahmedabad, INDIA www.facebook.com/Prasanth Sivakumar FOLLOW ME ON ACCESS MY RESOURCES IN SLIDESHARE prasanthperceptron CONTACT ME [email_address]](https://image.slidesharecdn.com/sequencealignments-completecoverage-110324014158-phpapp02/85/Sequence-alignments-complete-coverage-1-320.jpg)




![Global Alignment: Needleman-Wunsch Algorithm Matrix Fill Step G-G match score = +5 Si,j = MAX [0 + 5 , -4 + -4 , -4 + -4 ] = MAX [ 5 , -8 , -8 ] = 5 Confusing ? Diagonal + Match/Mismatch Score Left + Gap penalty Right + Gap penalty](https://image.slidesharecdn.com/sequencealignments-completecoverage-110324014158-phpapp02/85/Sequence-alignments-complete-coverage-7-320.jpg)
![Global Alignment: Needleman-Wunsch Algorithm G-A mismatch score = -3 Si,j = MAX [-4 + -3 , 5 + -4 , -8 + -4 ] = MAX [ -7 , 1 , -12 ] = 1](https://image.slidesharecdn.com/sequencealignments-completecoverage-110324014158-phpapp02/85/Sequence-alignments-complete-coverage-8-320.jpg)