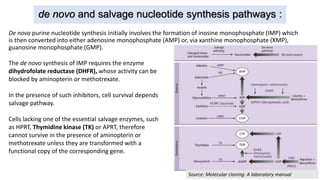
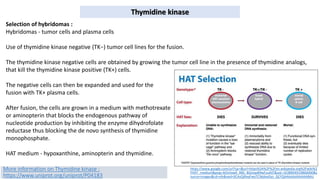
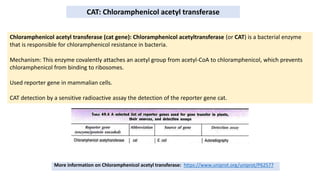
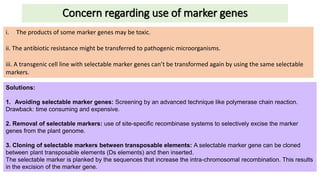
Thymidine kinase (TK), dihydrofolate reductase (dhfr), and chloramphenicol acetyl transferase (CAT) are examples of selectable marker genes used for animal cells. Marker genes help monitor transfection by allowing detection of whether the transgene was successfully transferred. Selectable marker genes enable transformed cells to survive selection conditions that kill non-transformed cells.