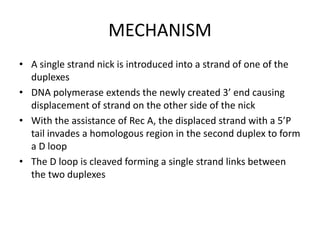
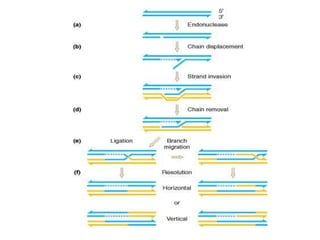
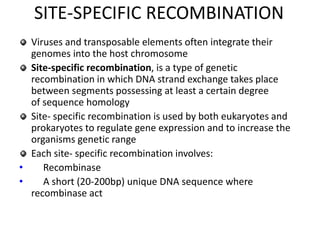
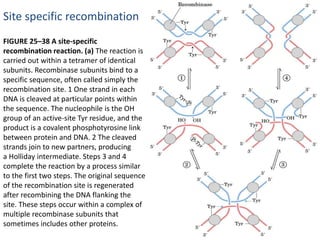
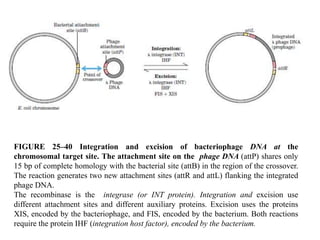
The document discusses genetic recombination and site-specific recombination. It describes the Meselson-Radding model of genetic recombination, which involves a single-strand nick that allows DNA polymerase to extend the 3' end and displace the other strand, forming a D loop structure. Site-specific recombination involves recombinases cutting DNA at specific recognition sequences and rejoining the strands to form a Holliday junction intermediate. Examples discussed include bacteriophage lambda integration into E. coli DNA, which is mediated by lambda integrase recombining attP and attB sites.