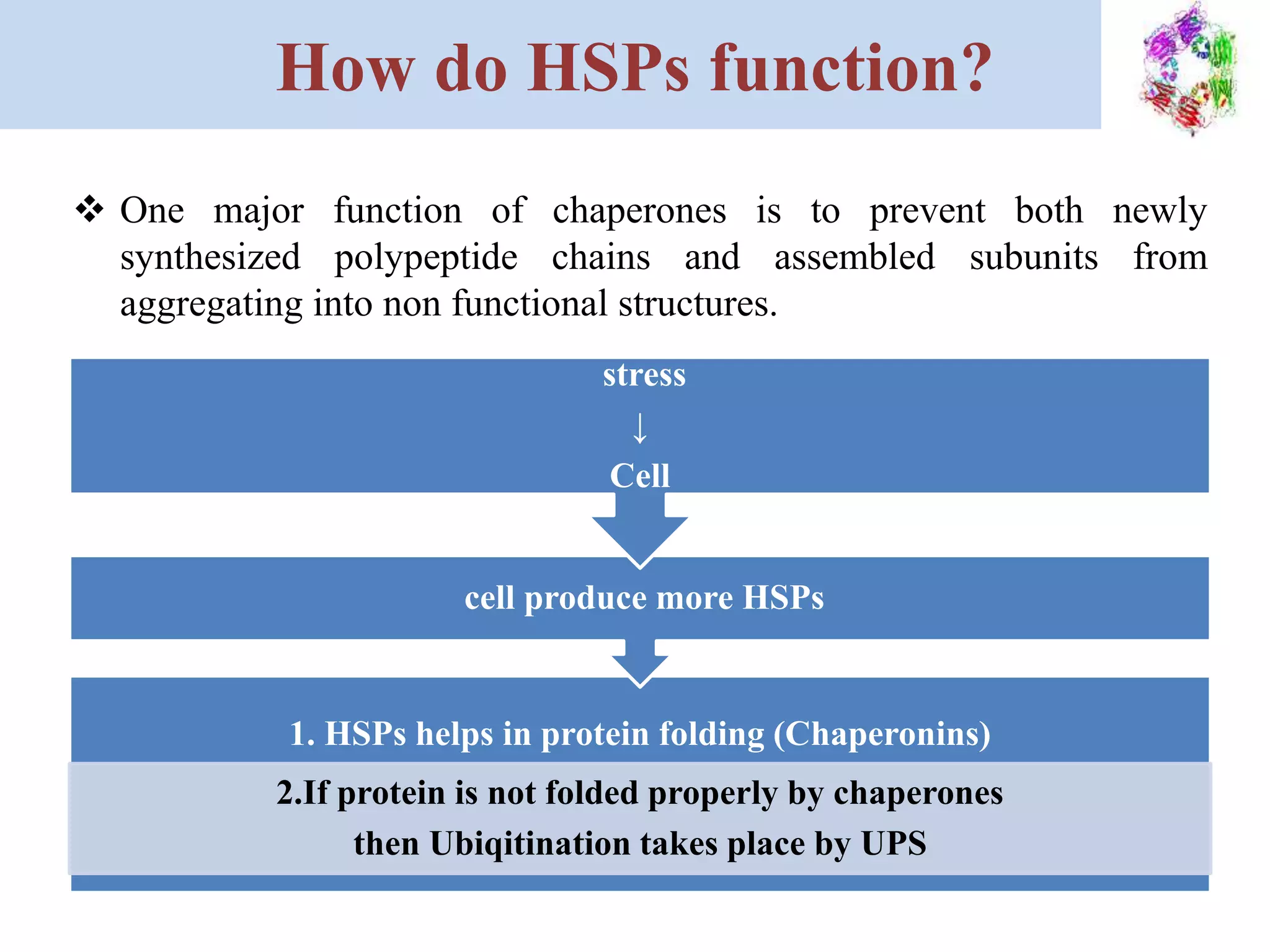
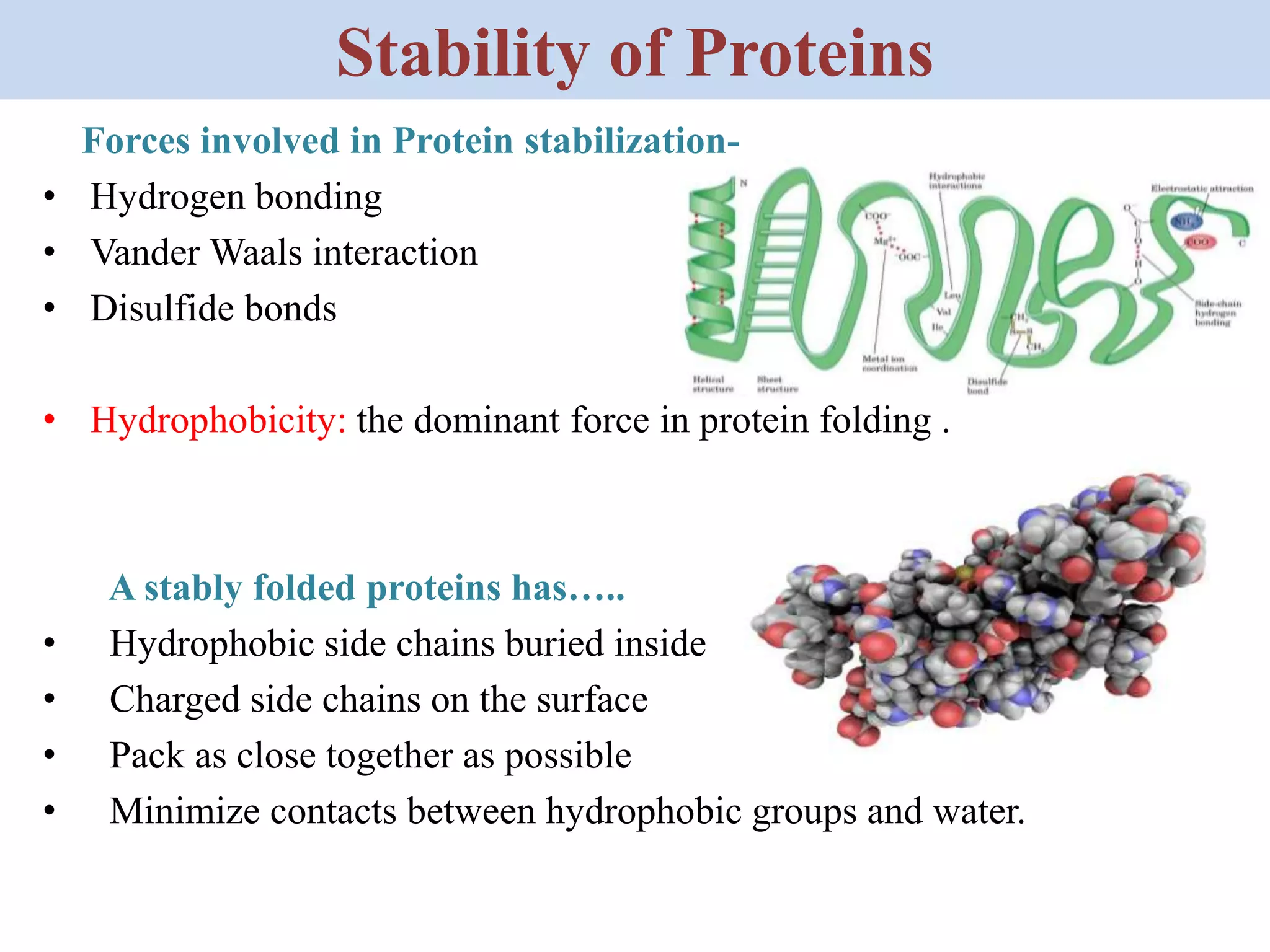
Heat shock proteins (HSPs) are highly conserved proteins found in all organisms that help proteins properly fold and prevent protein aggregation. HSPs are produced when cells experience stress like heat, cold, radiation, or toxins. Ritossa discovered HSPs in fruit flies in 1960. There are several classes of HSPs classified by molecular weight that assist protein folding through different mechanisms. Misfolded proteins are targeted for degradation by ubiquitination and the proteasome to maintain protein quality control, but HSPs can also help refold proteins to prevent this outcome. HSPs play an essential role in protein homeostasis.