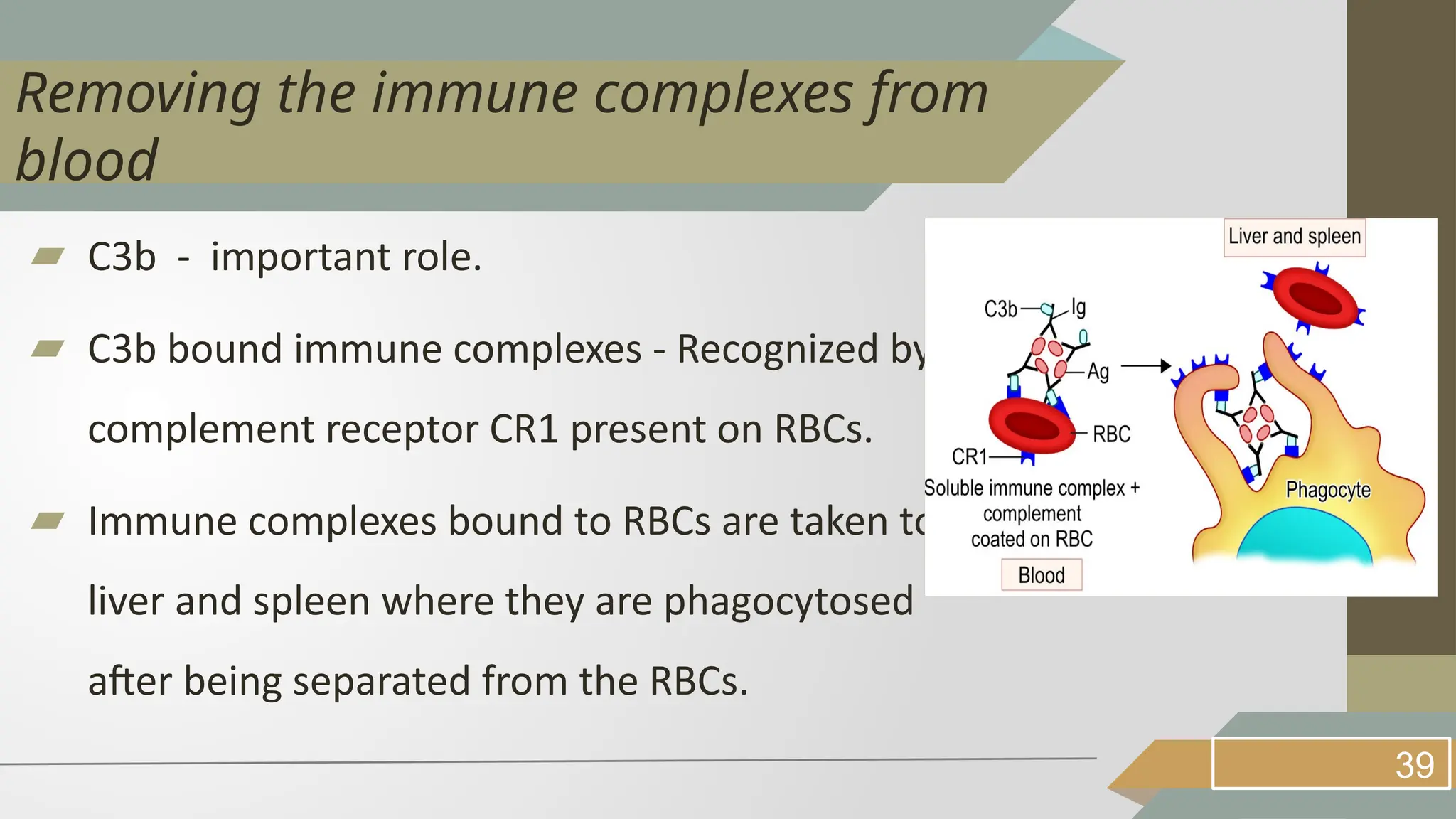
The complement system consists of about 30 serum proteins that are inactive in their normal state and play a crucial role in augmenting immune responses once activated. It includes classical, alternative, and lectin pathways for activation, each with distinct initiation processes but similar downstream effects leading to cell lysis through the formation of the membrane attack complex (MAC). Additionally, various mechanisms exist for microbial evasion of the complement system, and regulatory proteins are in place to moderate complement activity and prevent damage to host cells.