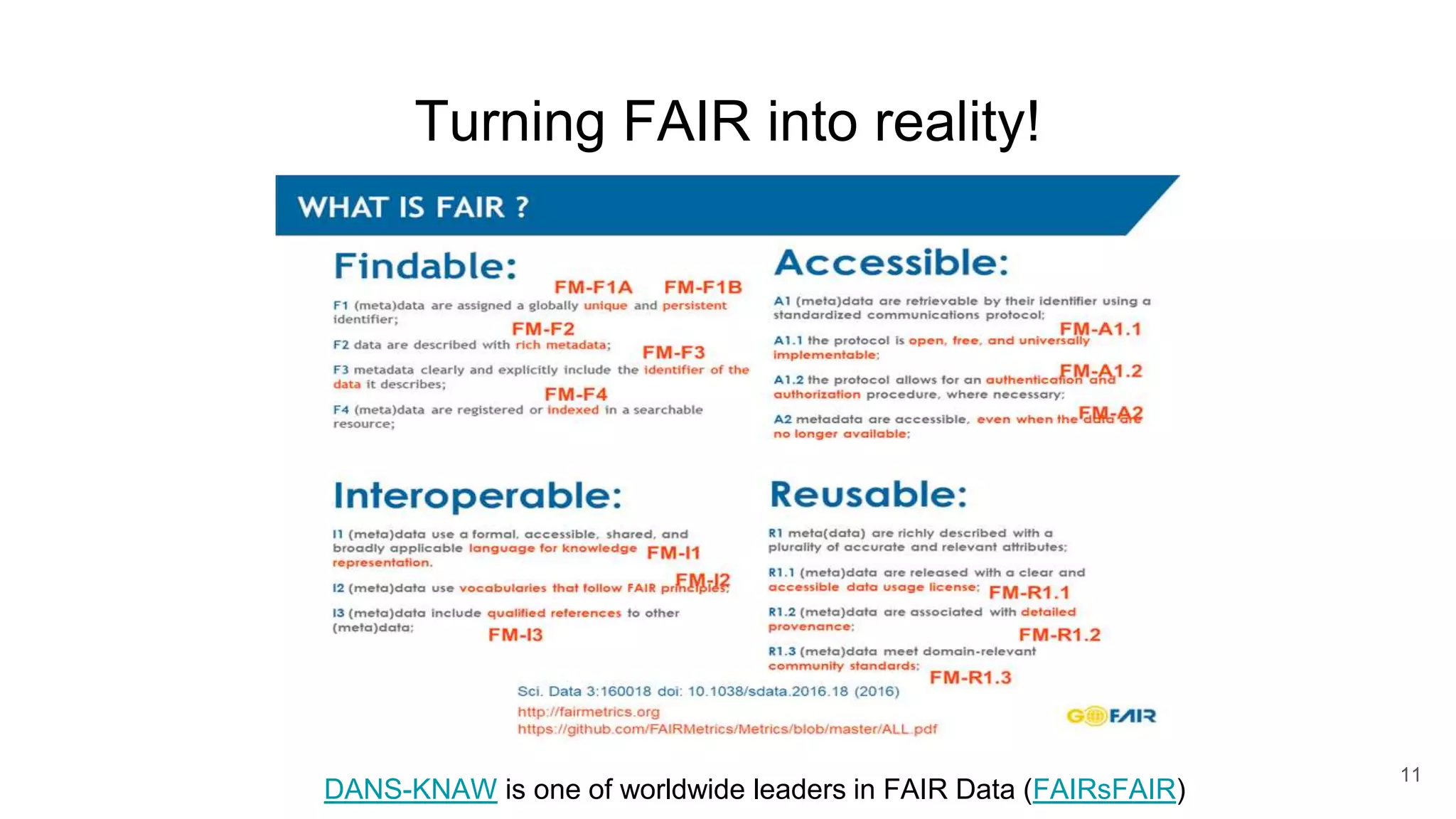
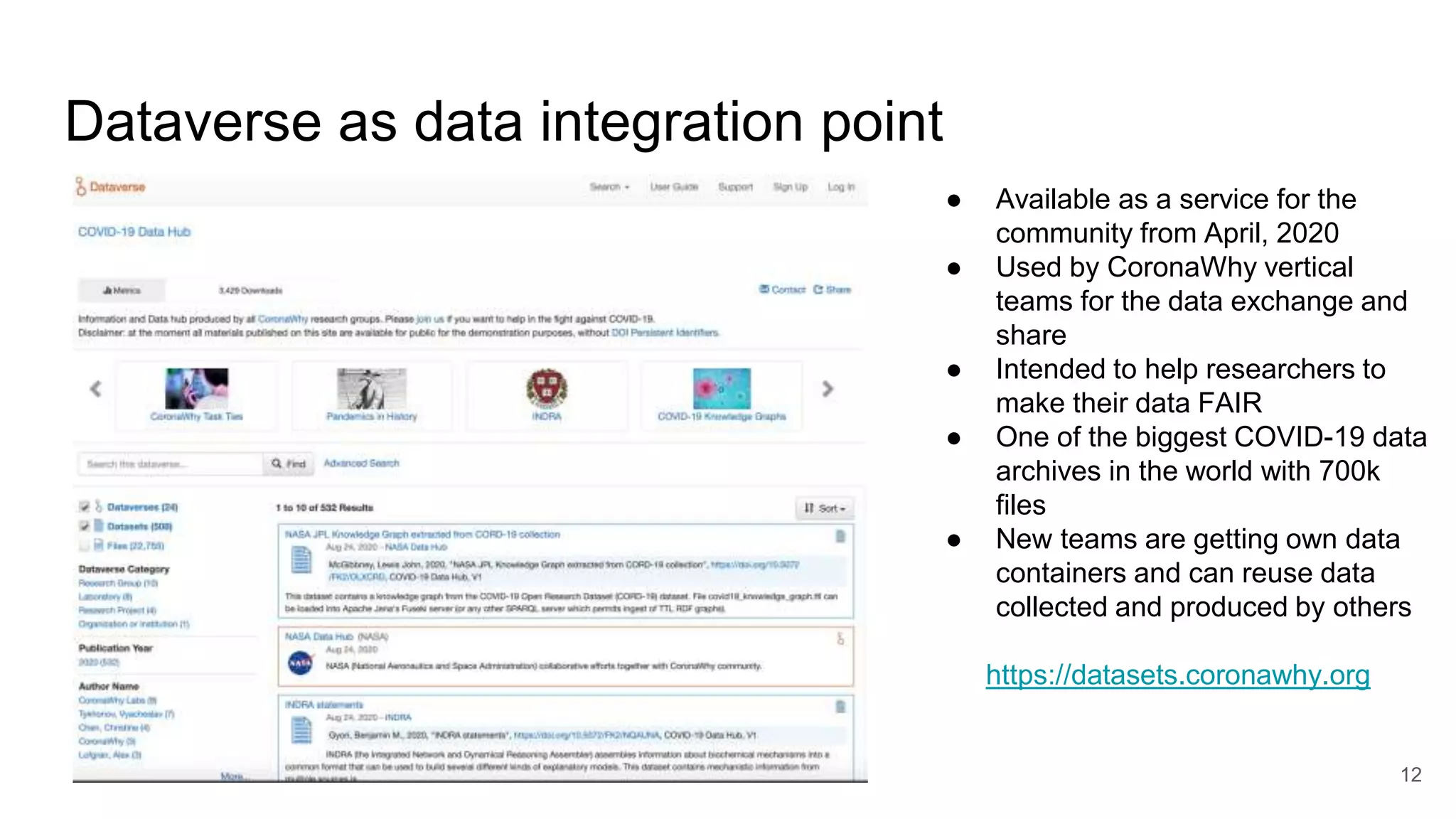
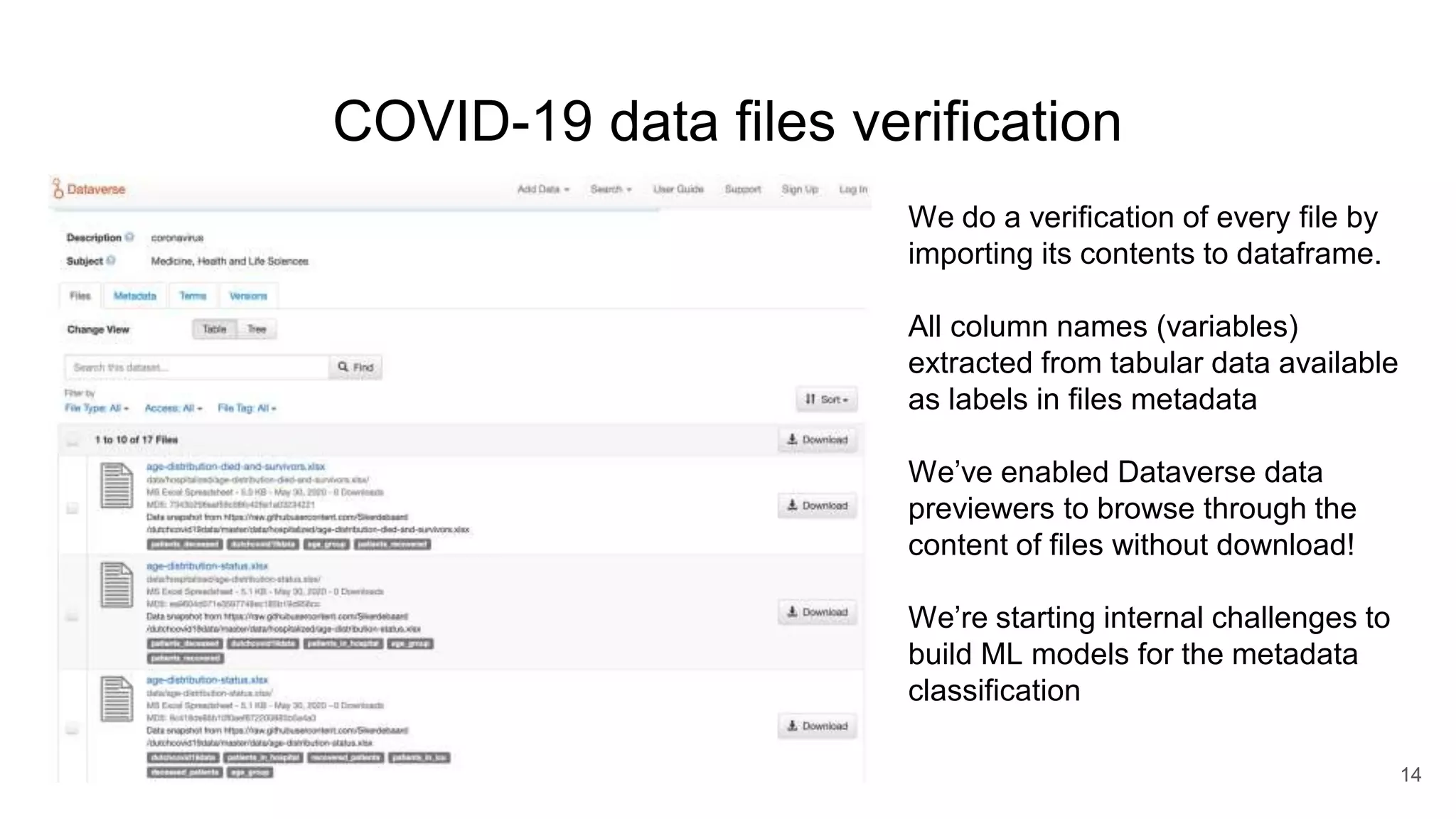
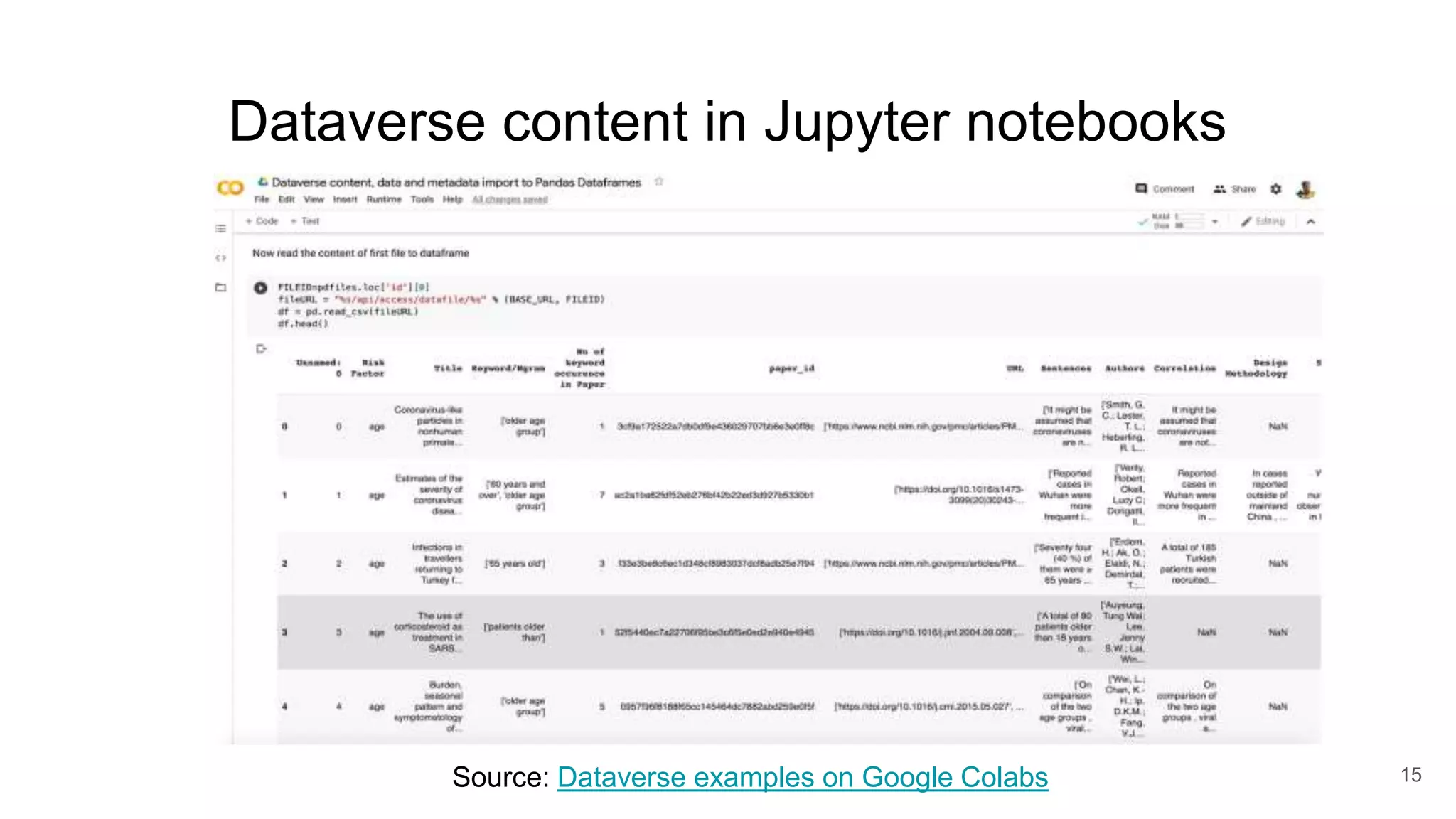
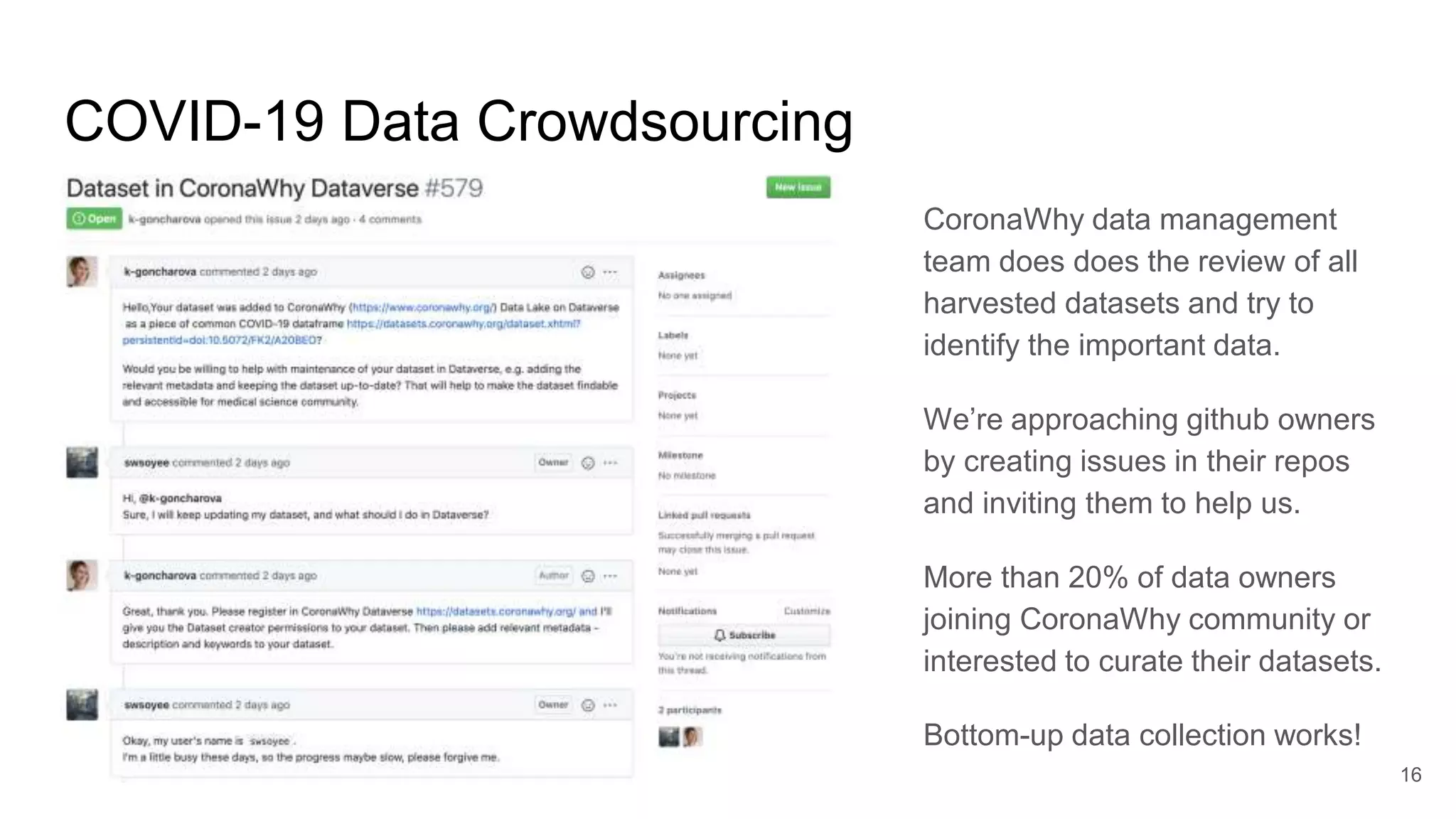
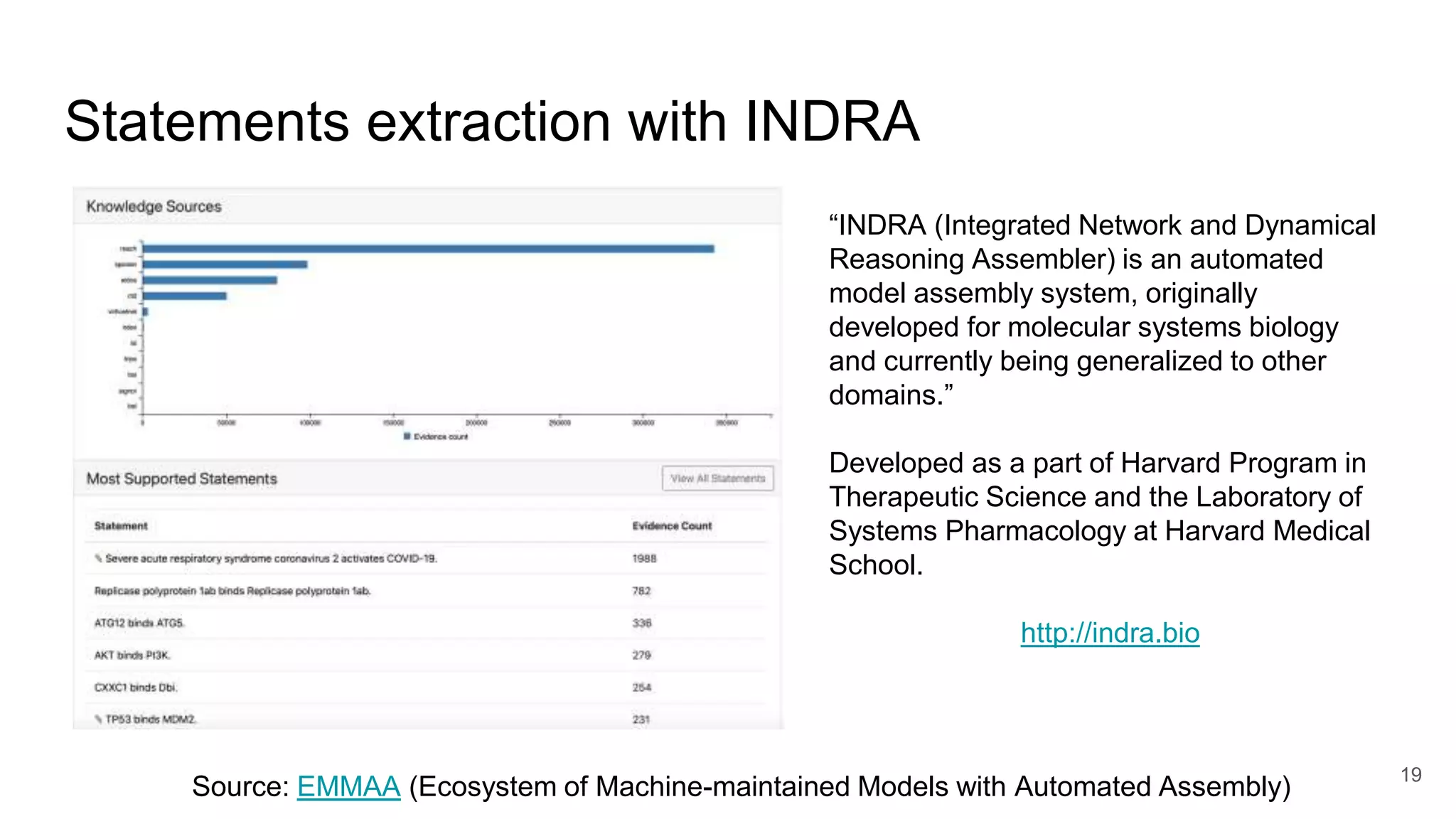
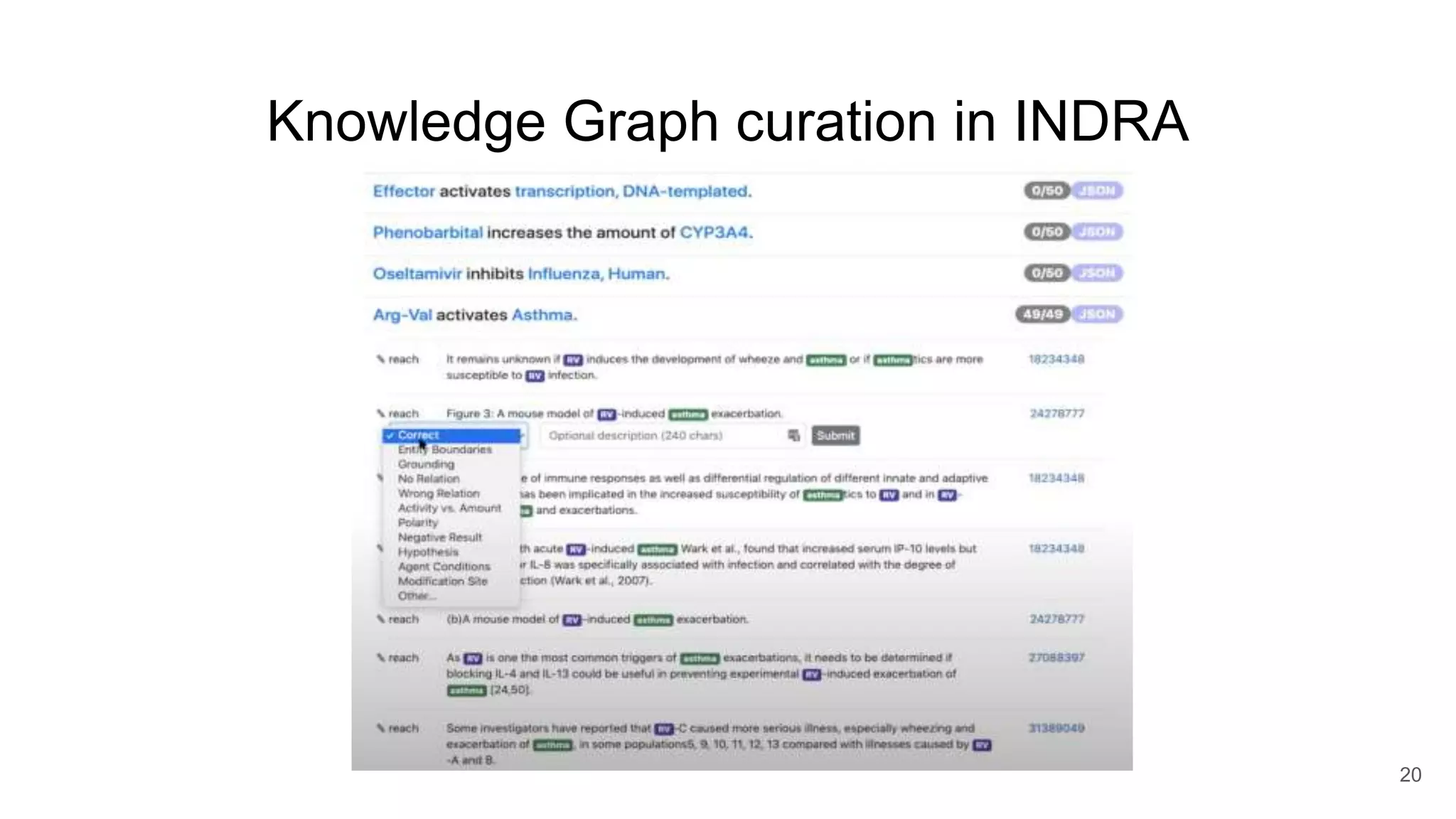
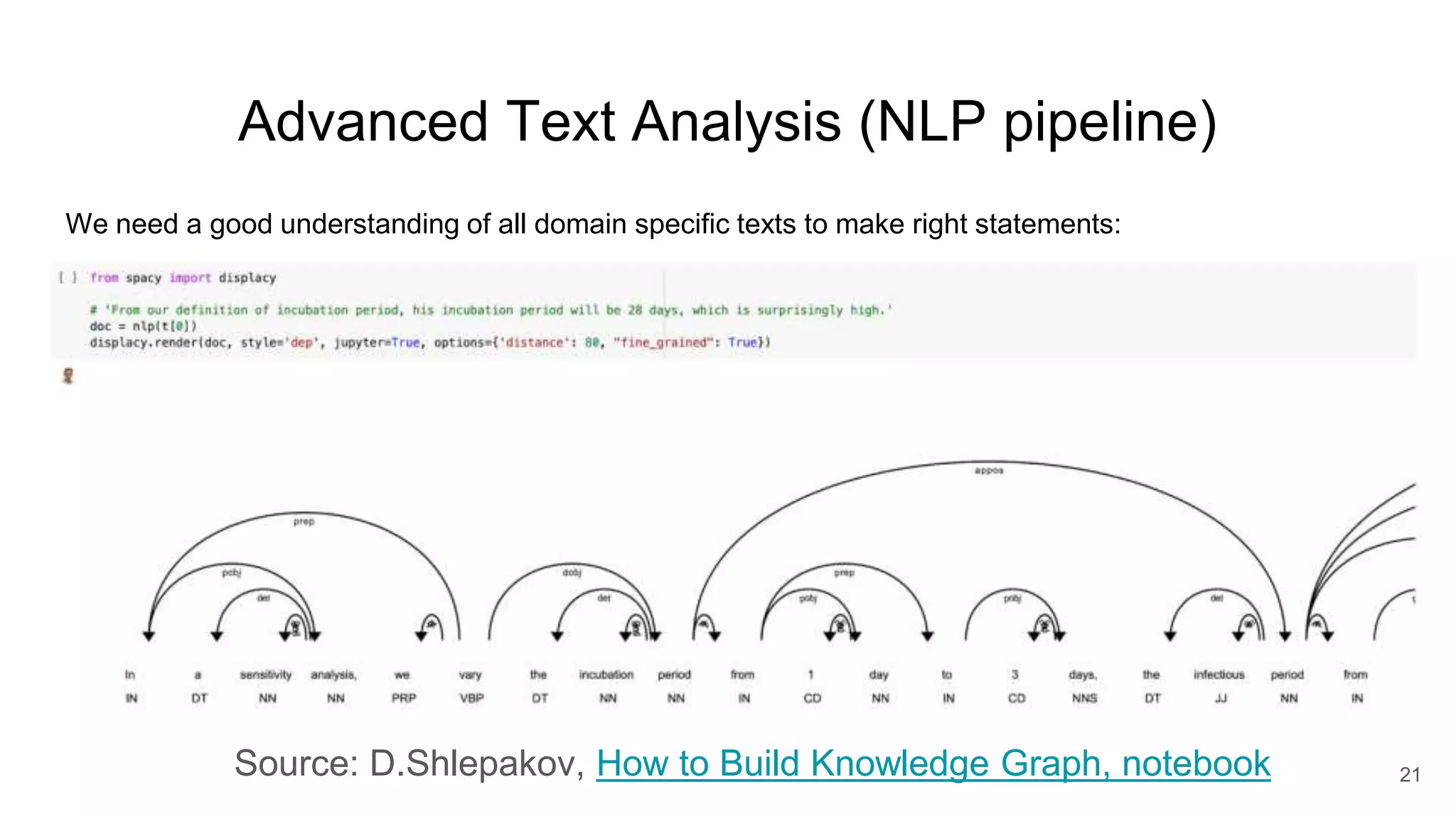
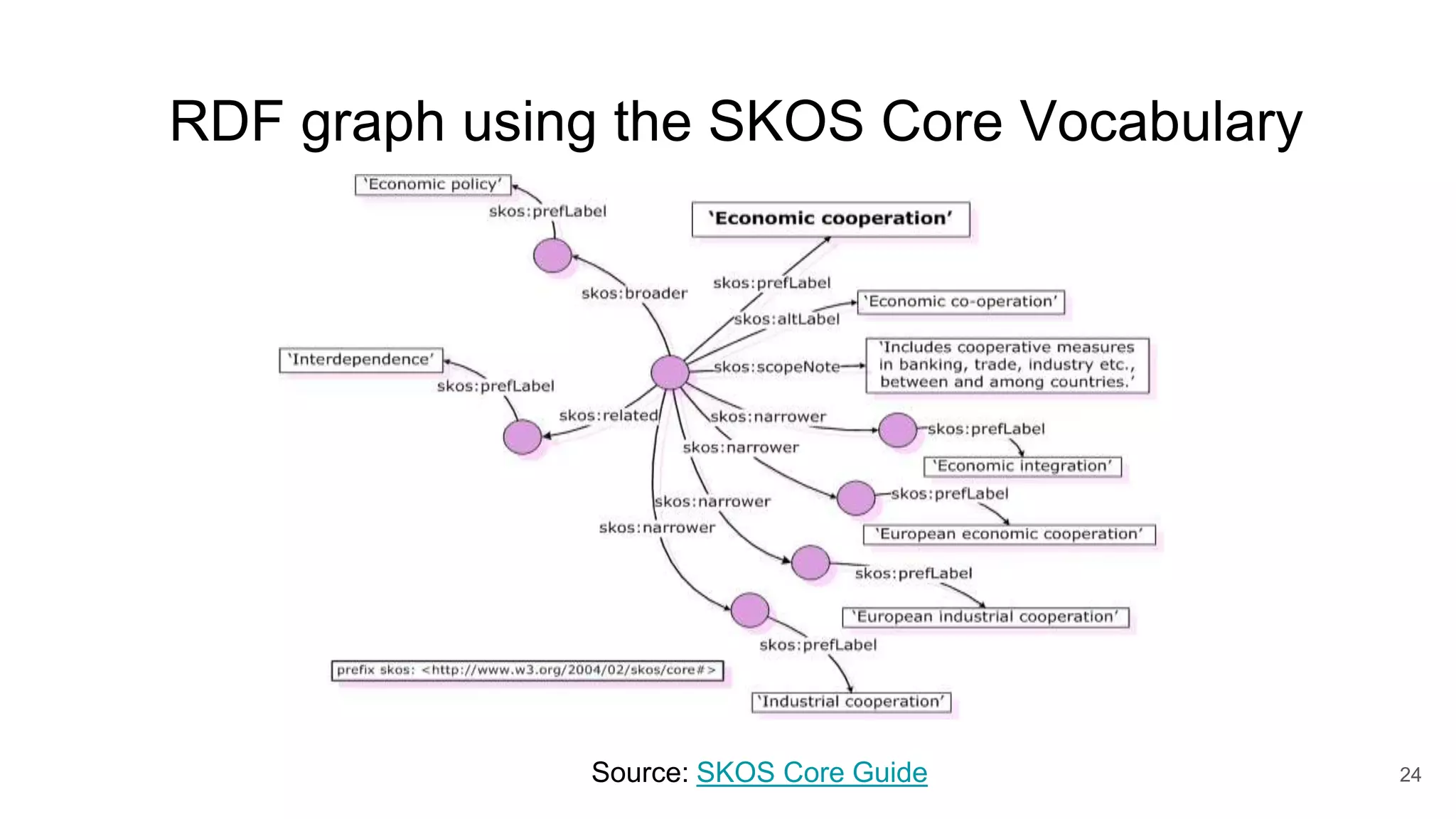
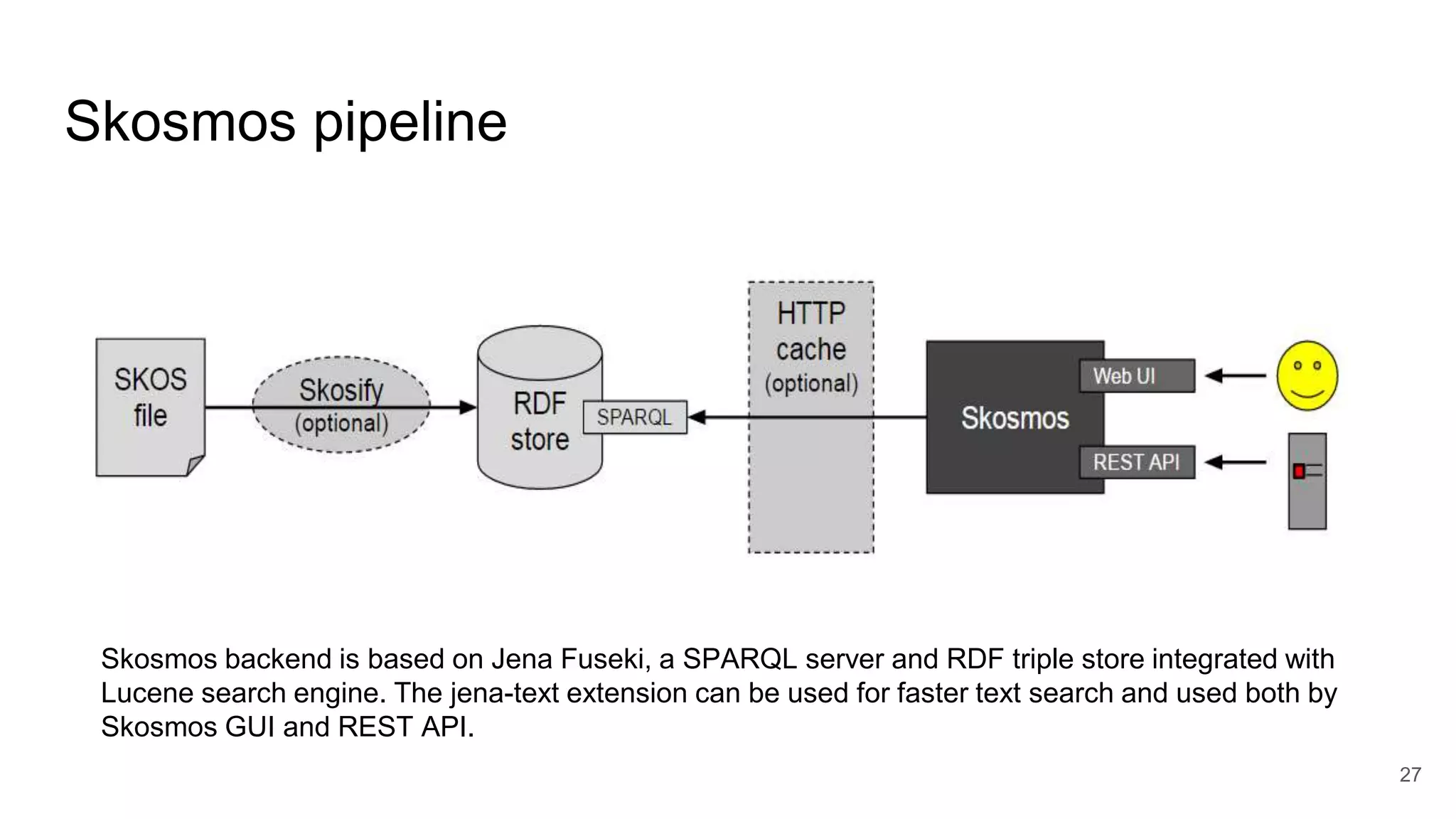
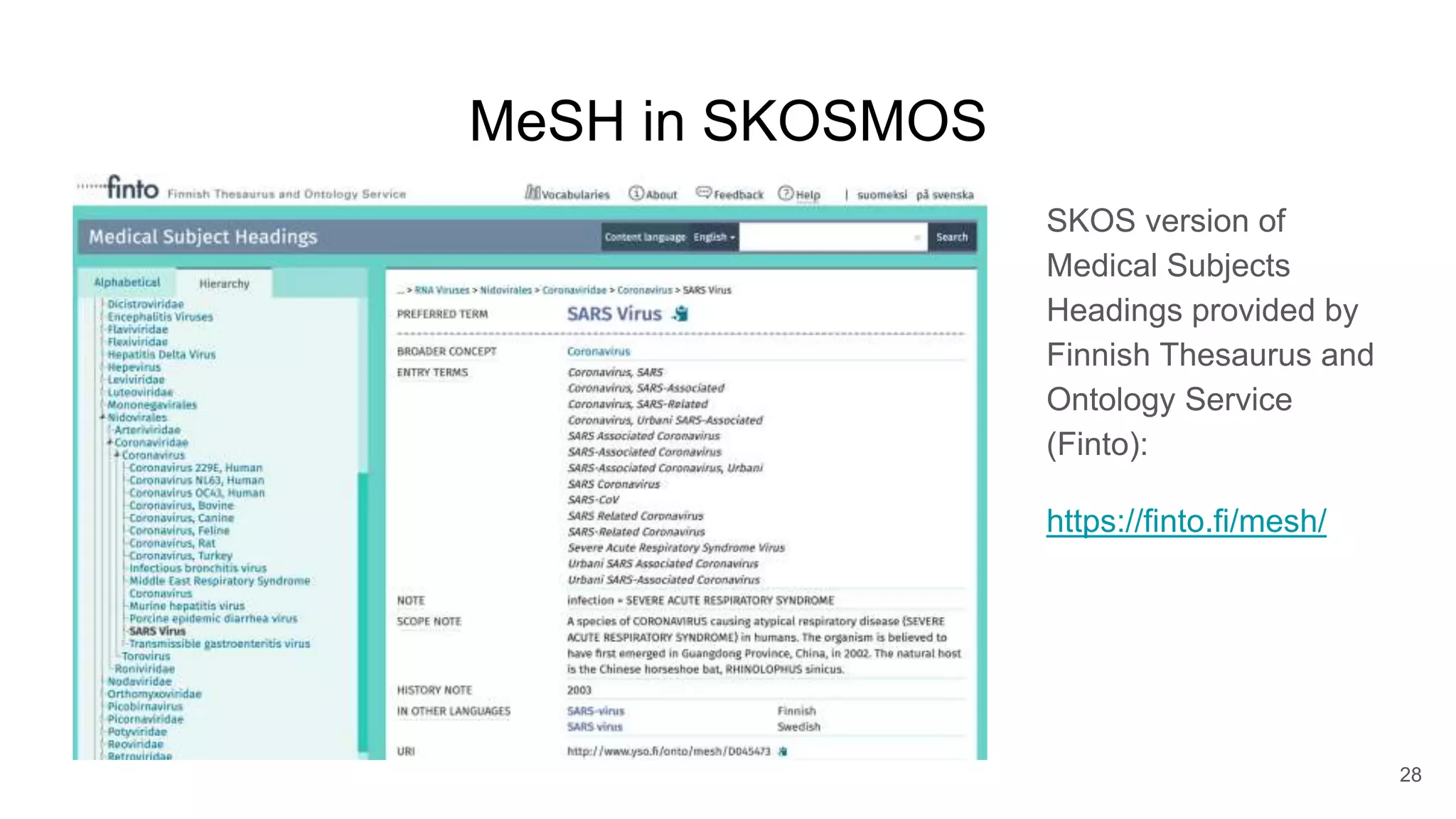
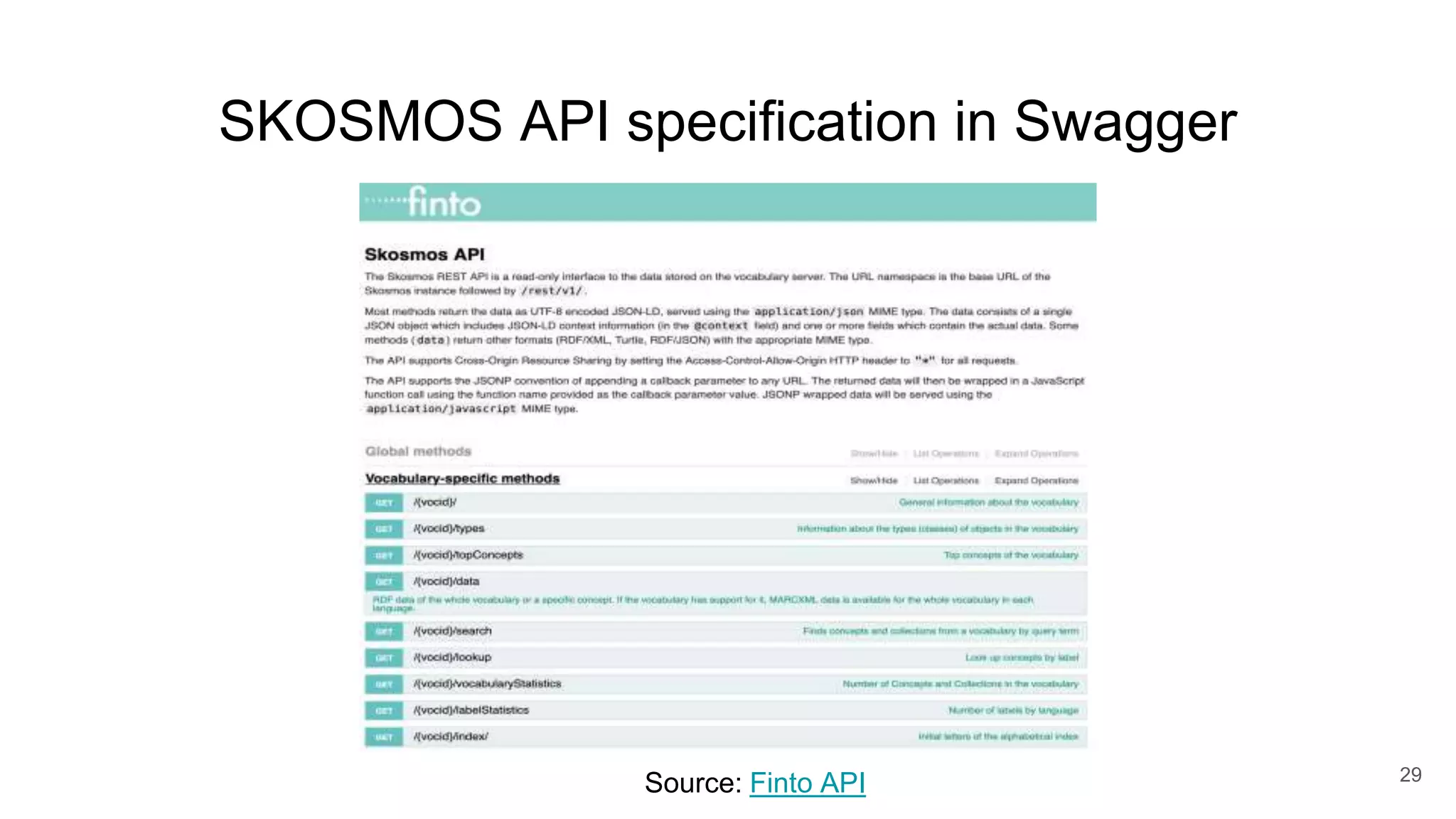
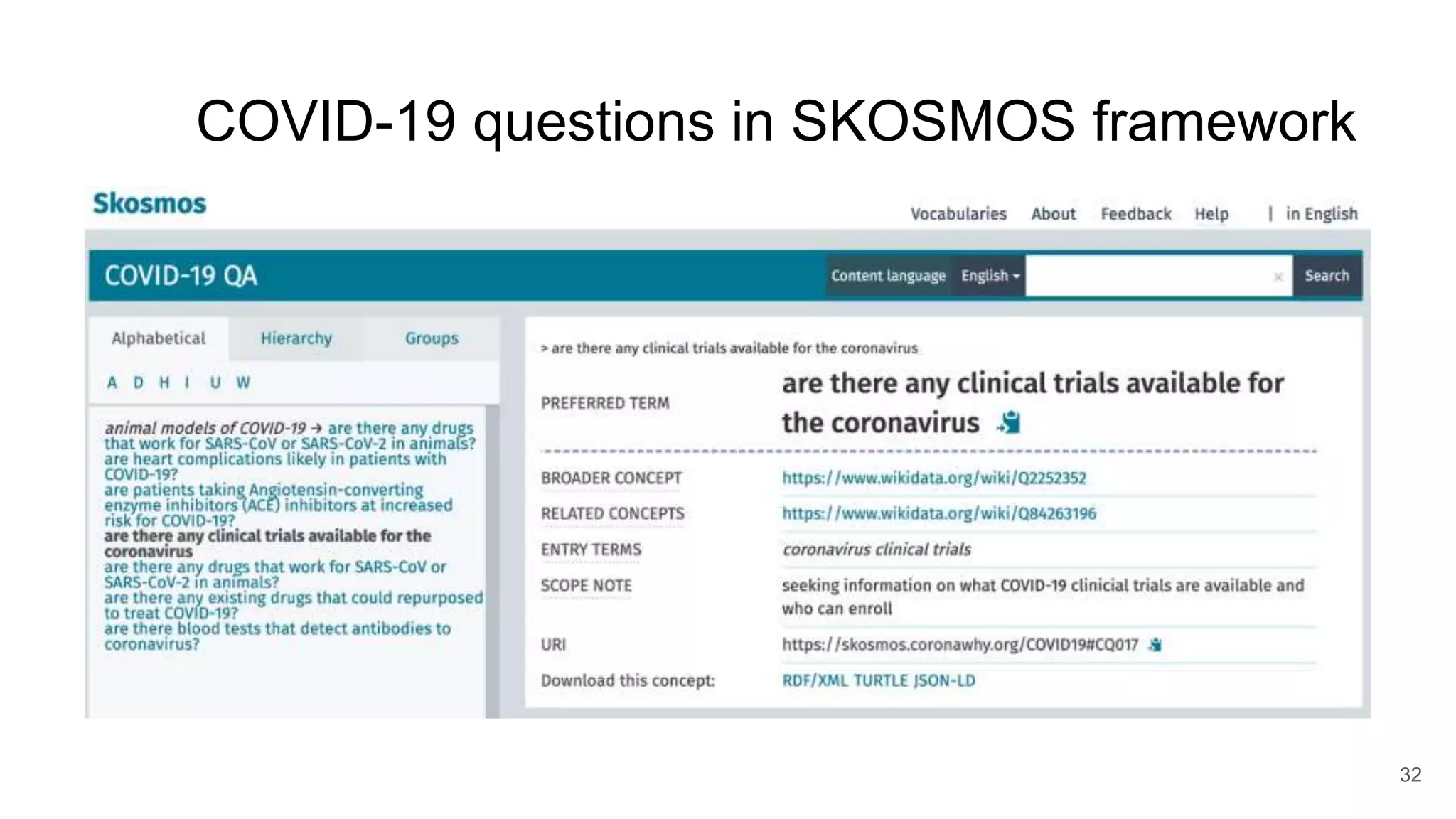
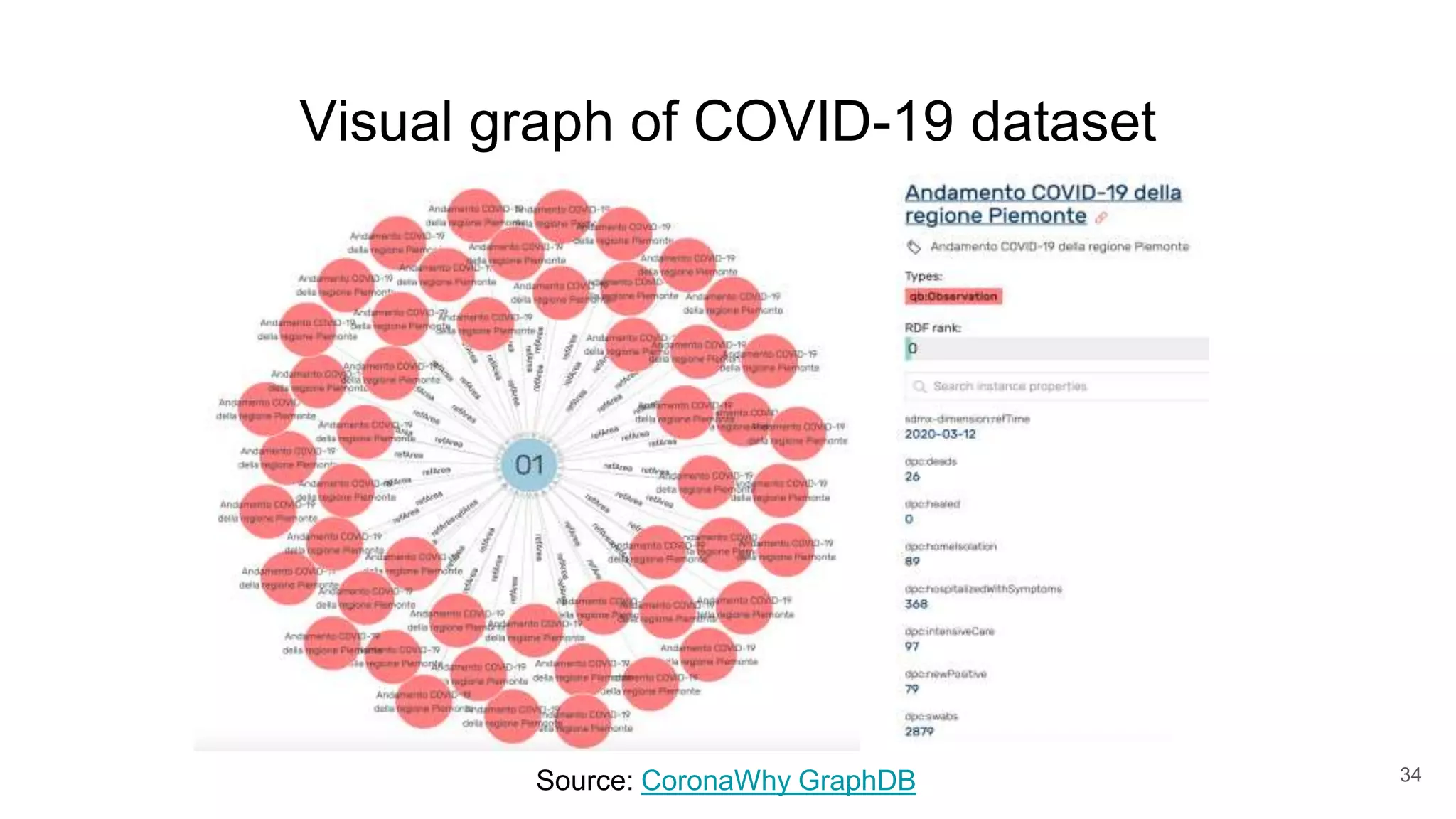
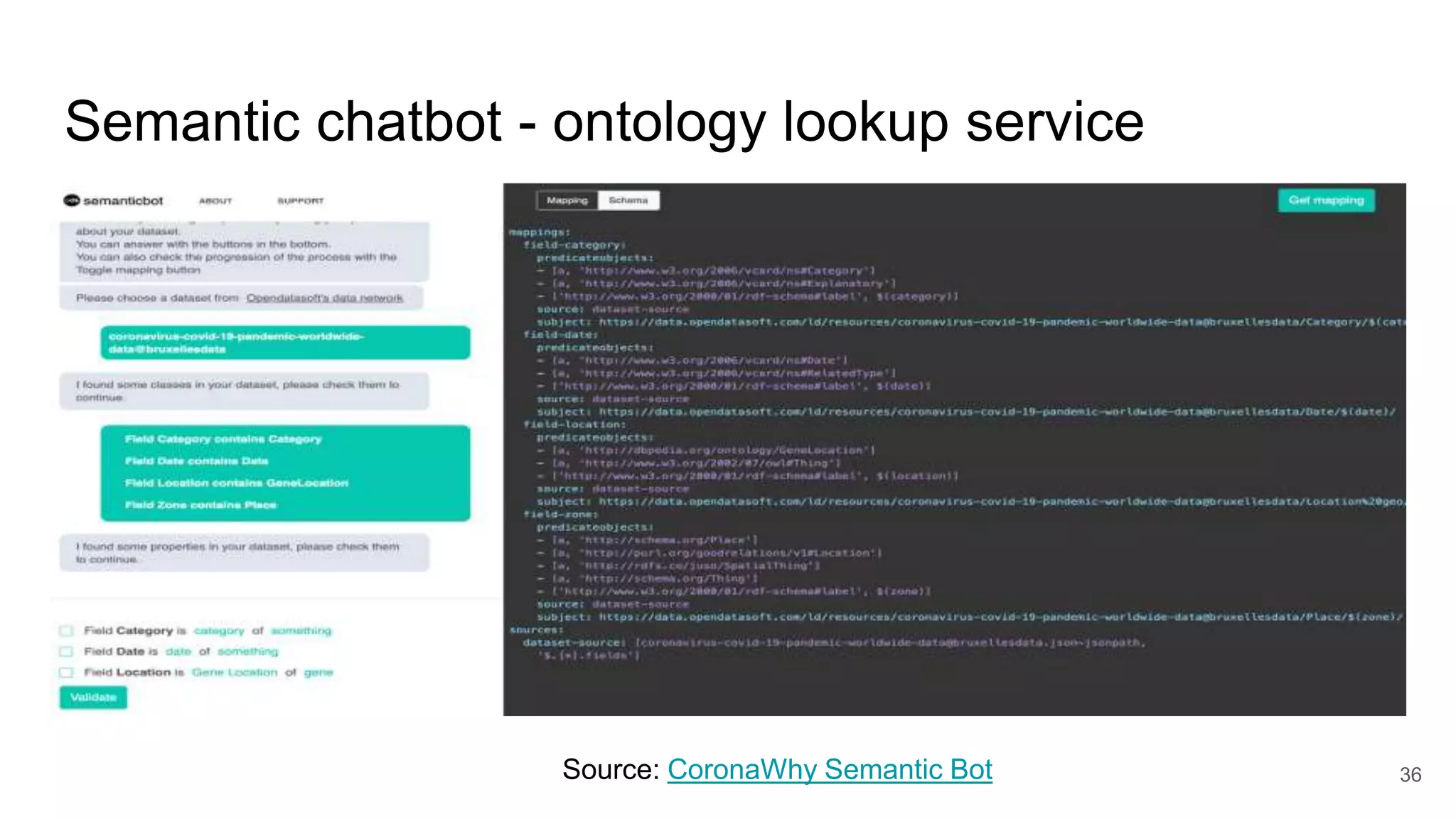
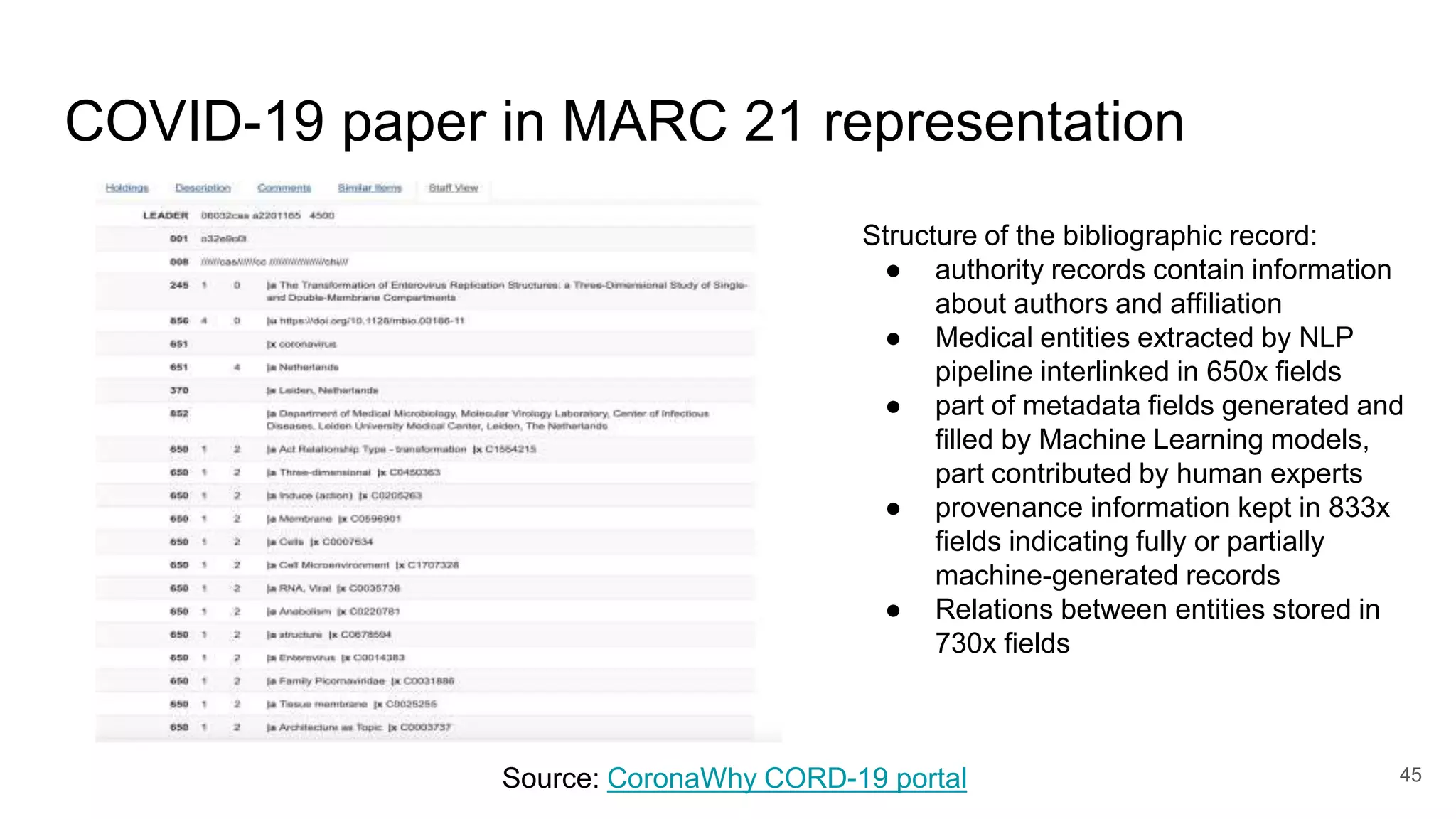
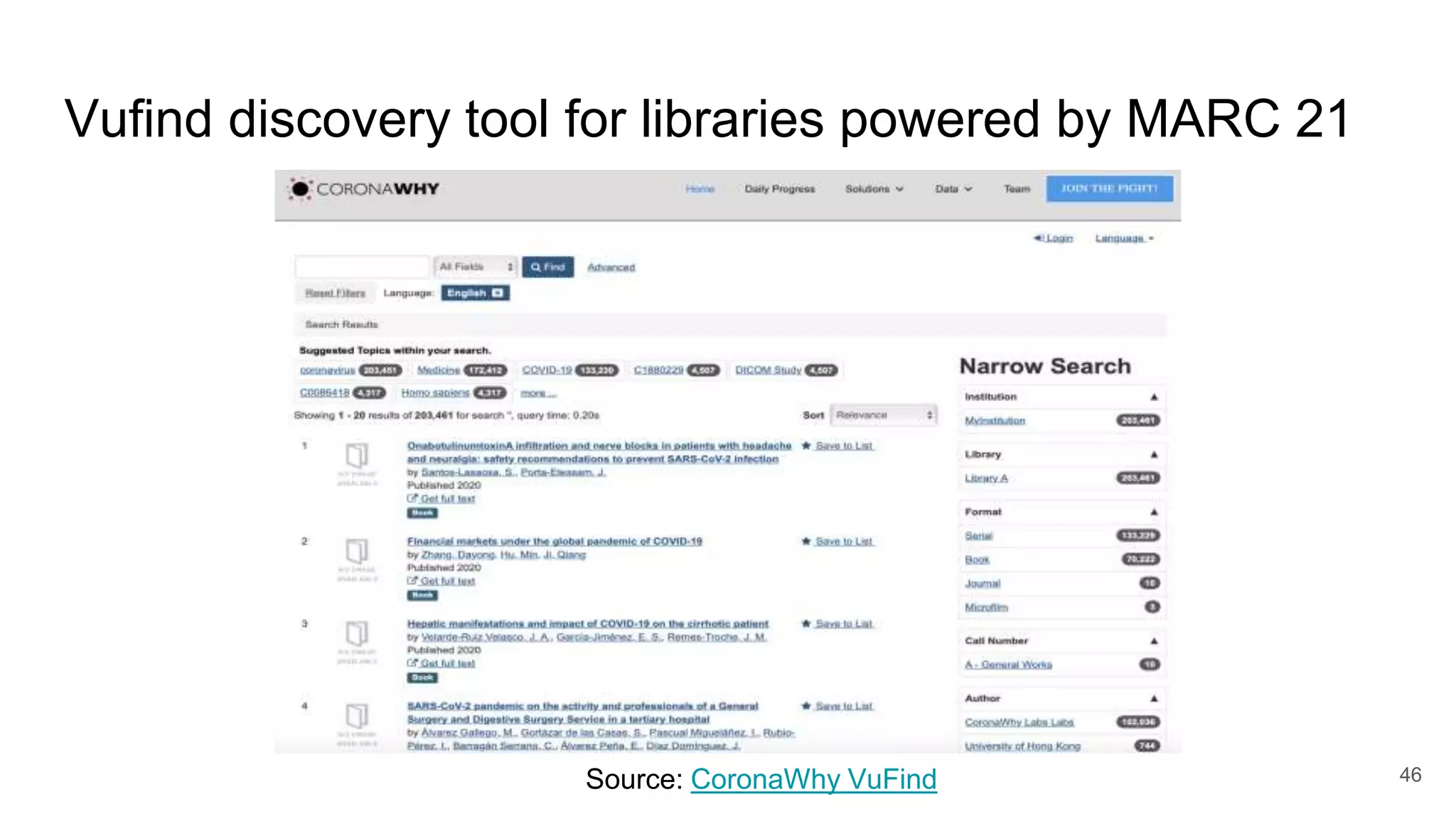
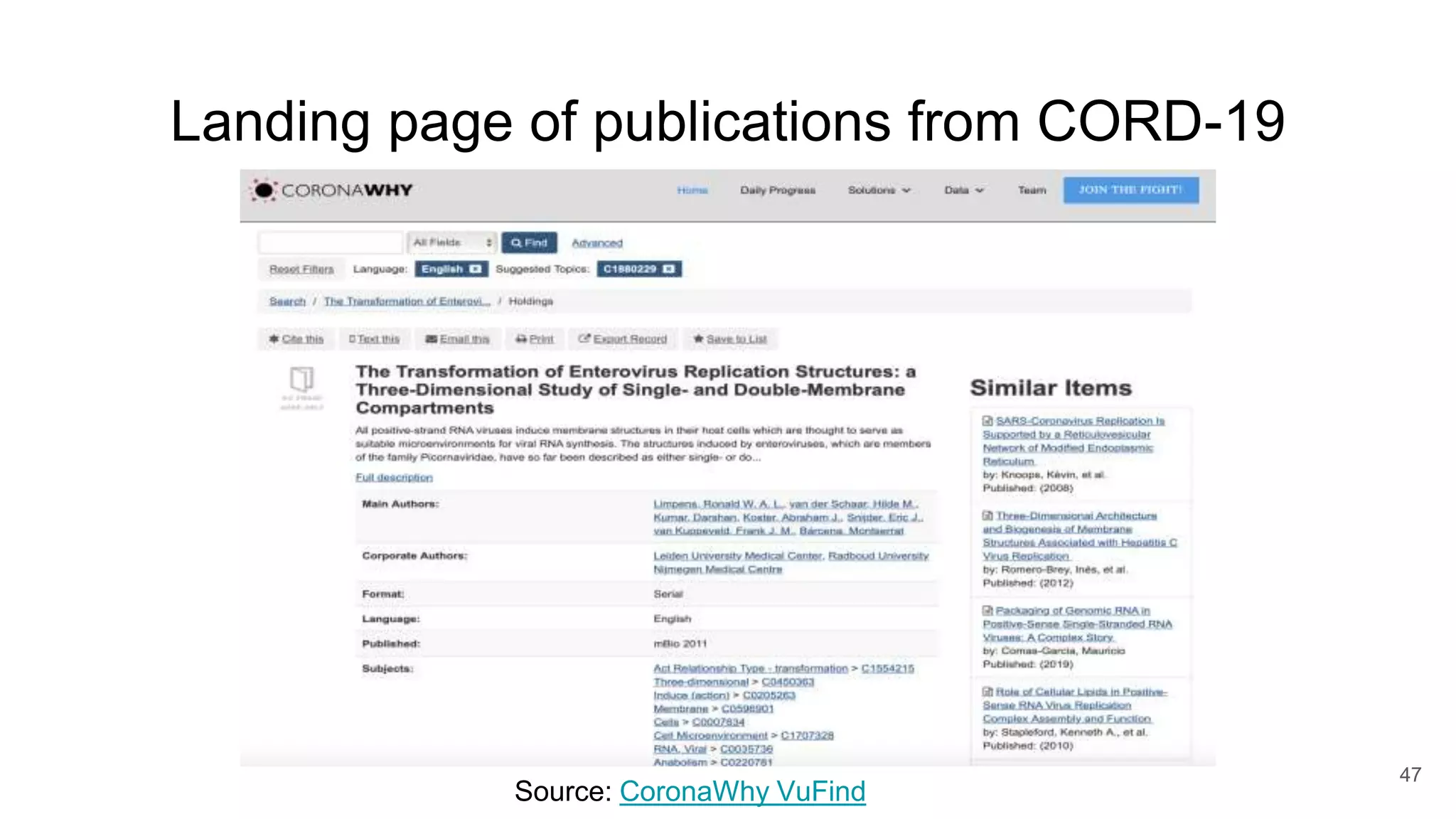
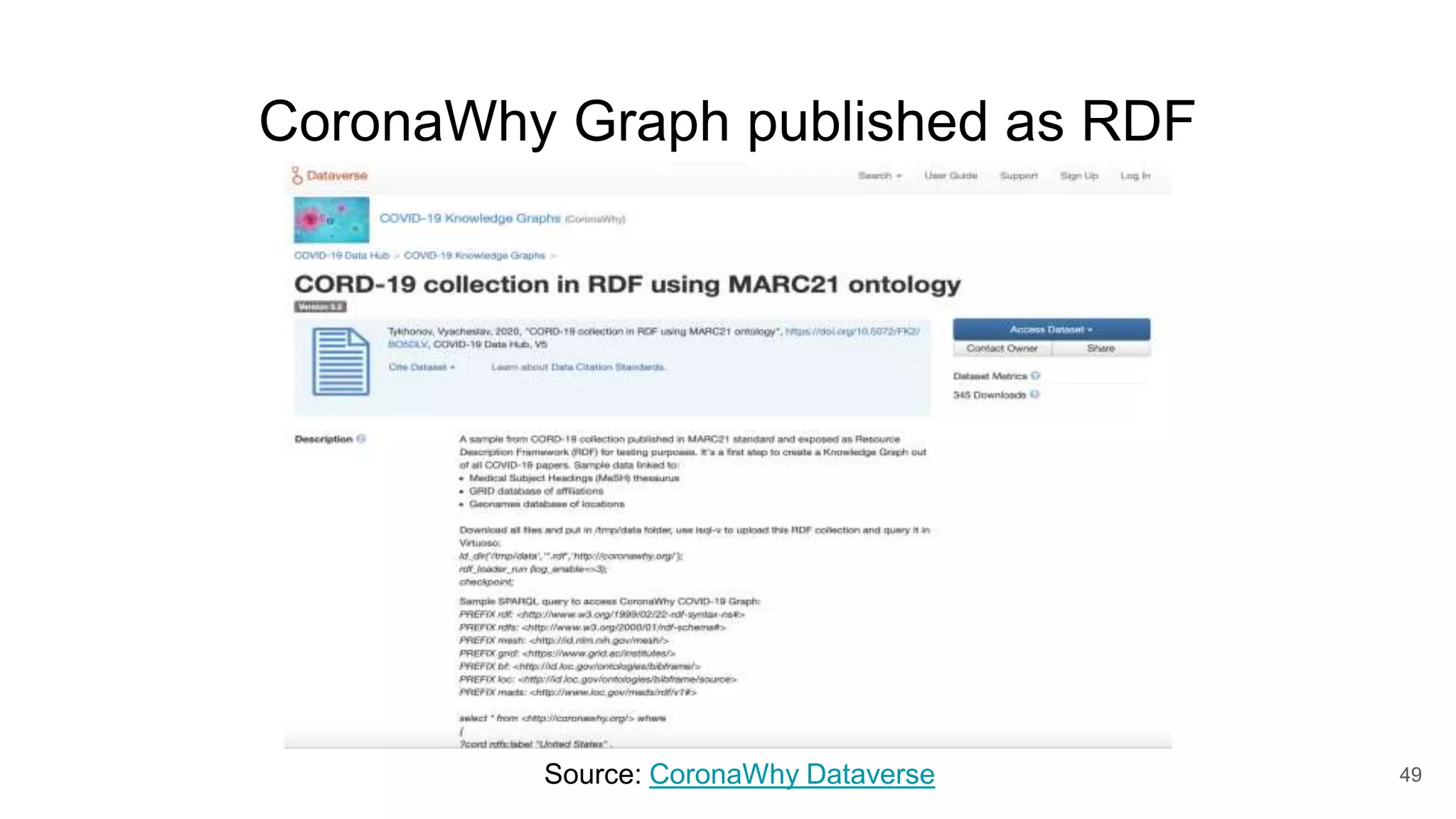
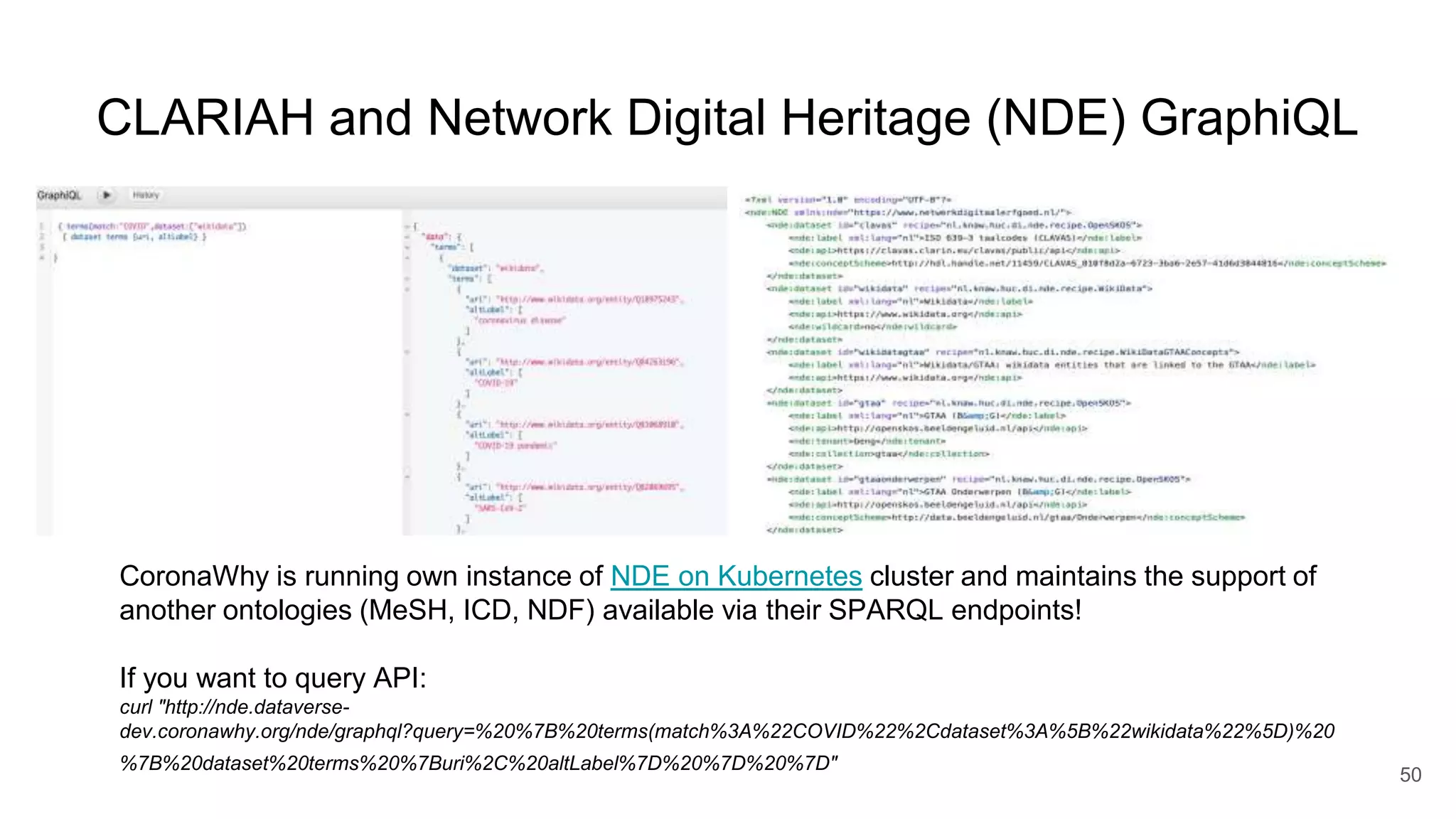
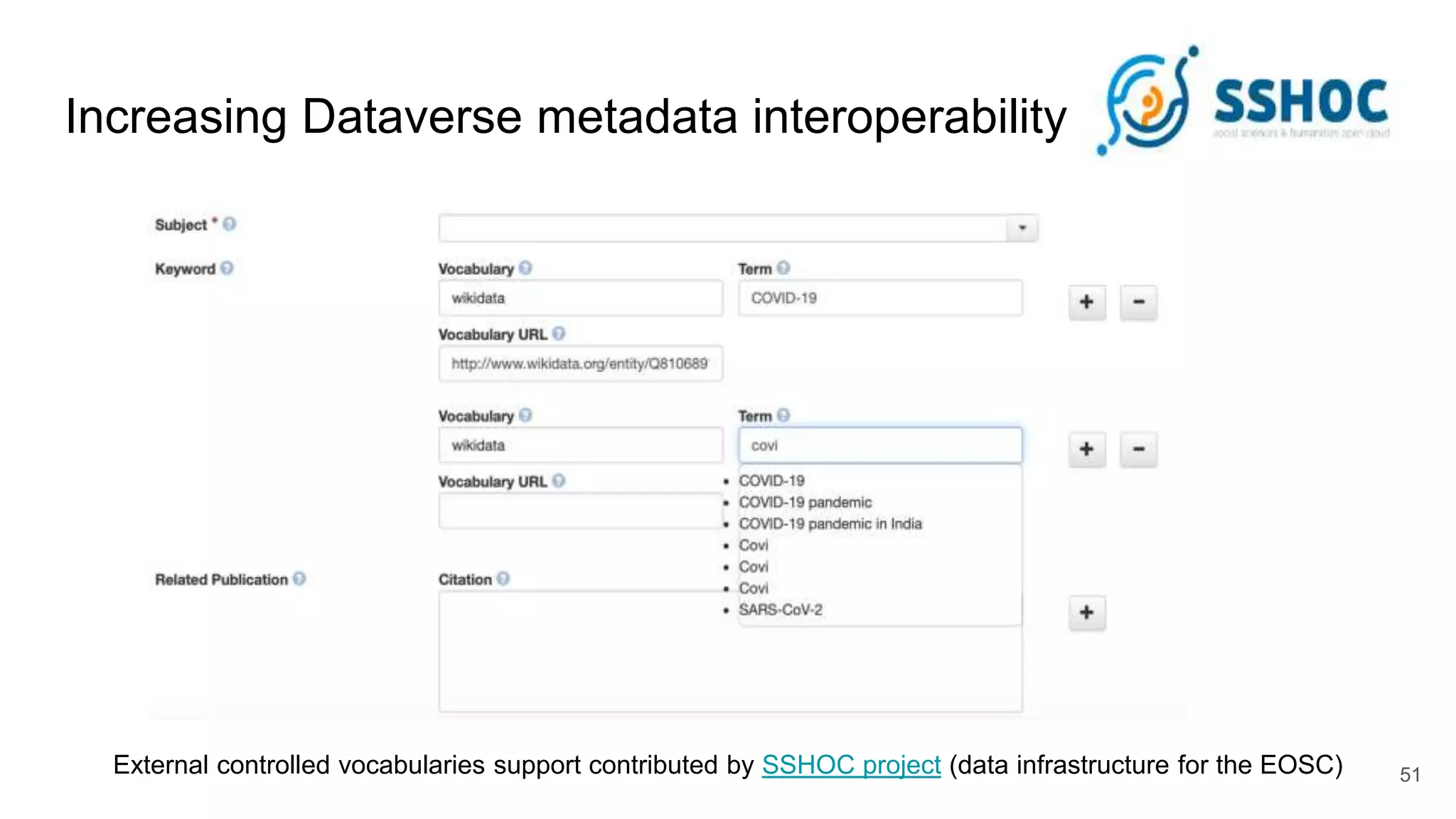
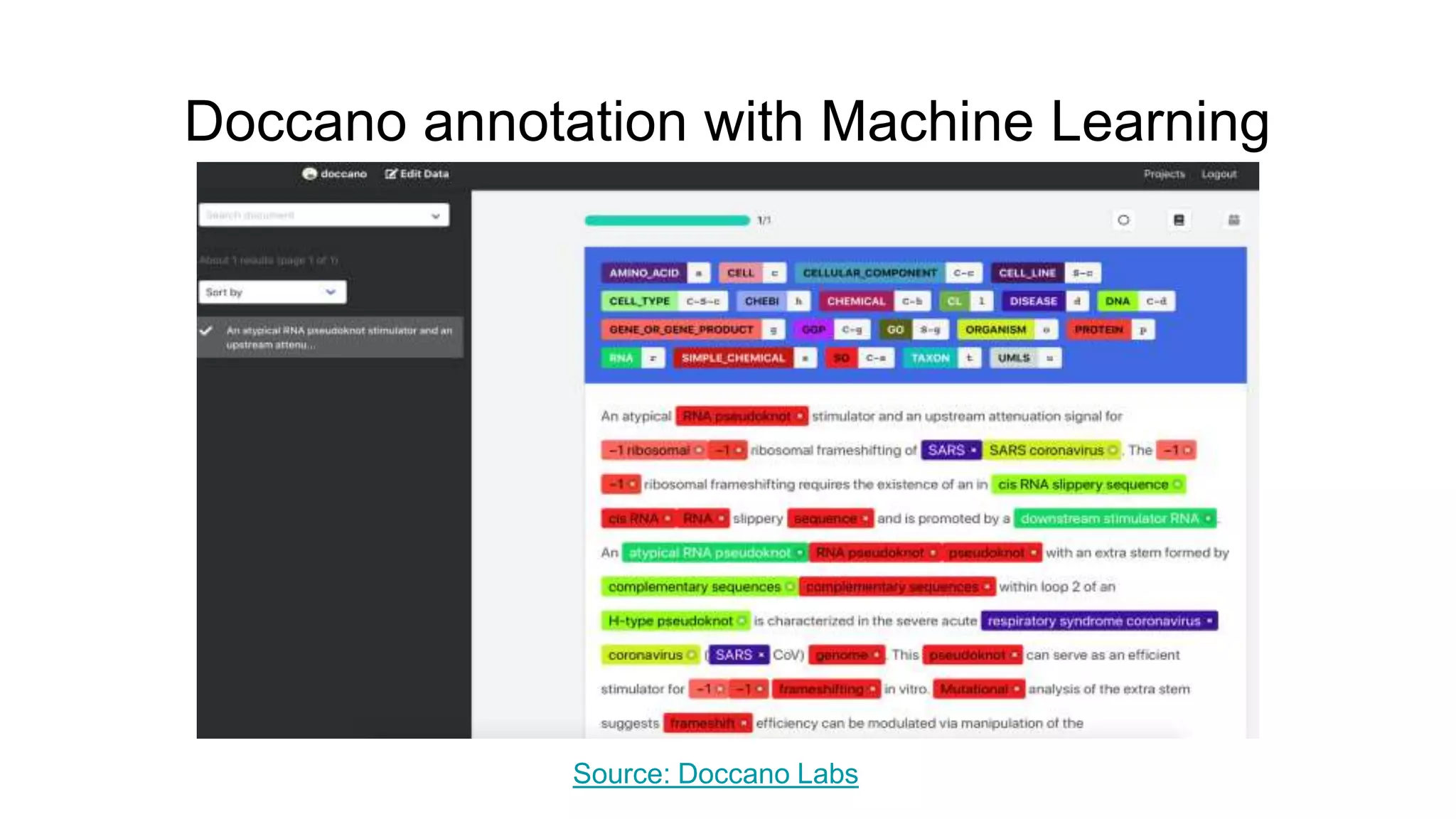
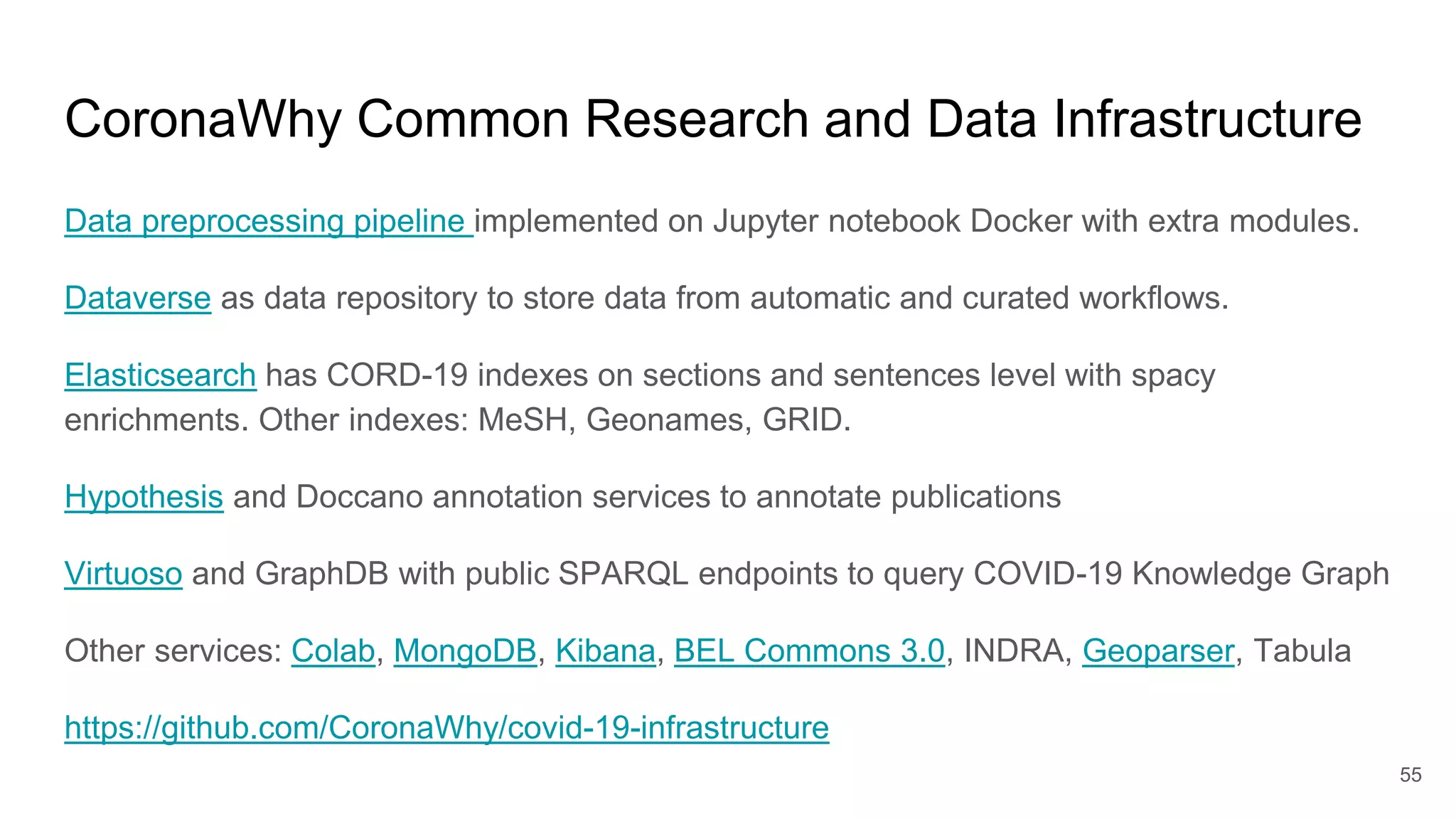
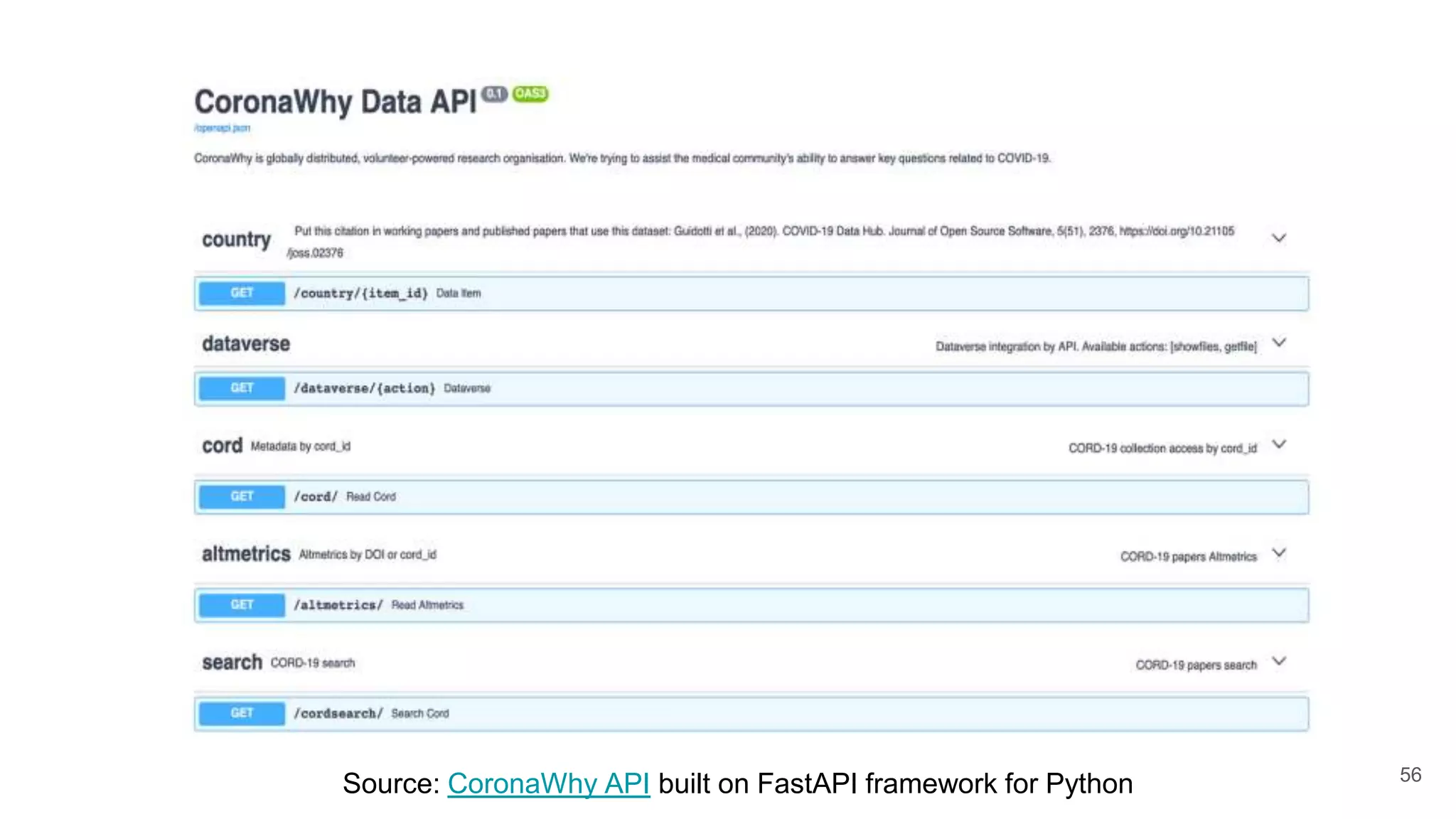
The document discusses the Coronawhy initiative, which aims to facilitate COVID-19 research through an extensive open research dataset and a collaborative knowledge graph. It highlights various tasks, partnerships, and technologies used for data integration, analysis, and visualization, with a focus on making research data fair and accessible. Additionally, it presents challenges in data interoperability and highlights the importance of controlled vocabularies and ontologies in enhancing research opportunities.