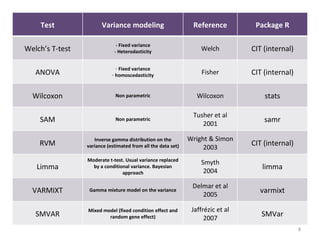
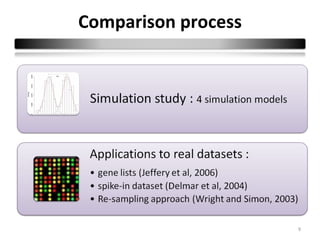
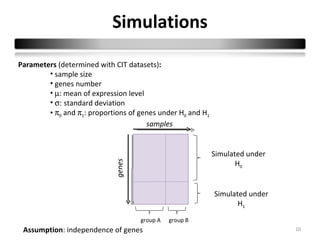
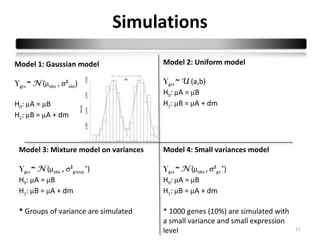
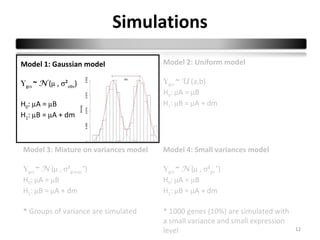
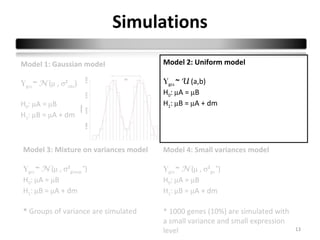
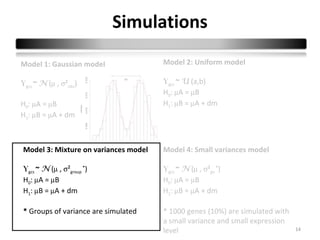
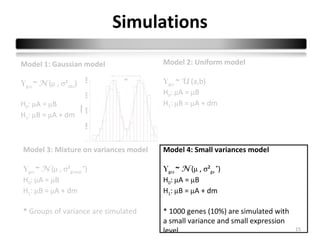
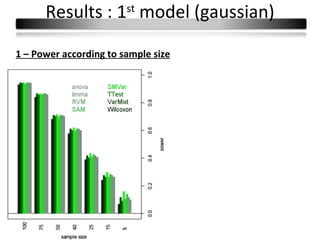
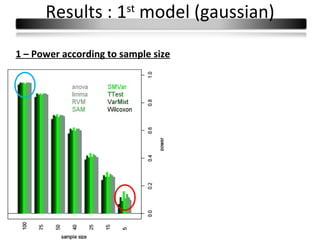
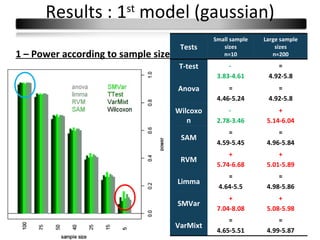
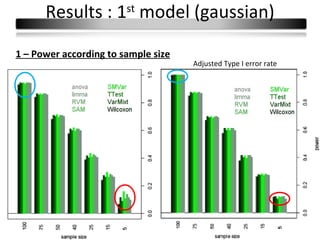
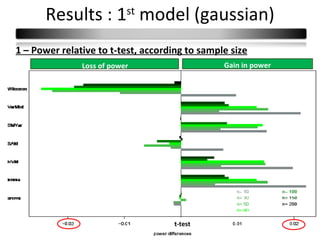
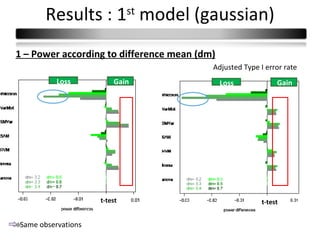
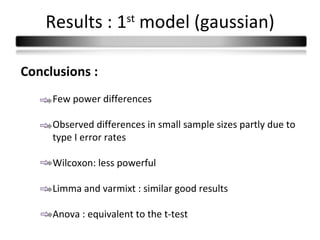
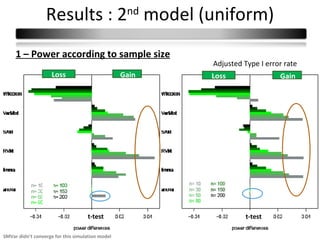
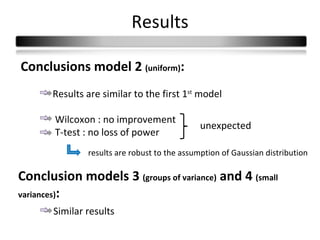
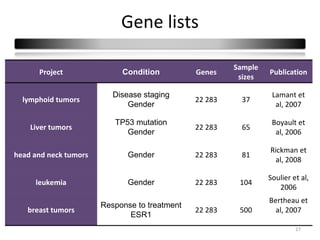
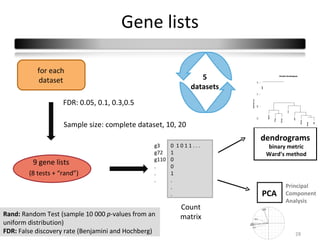
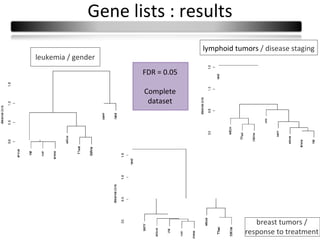
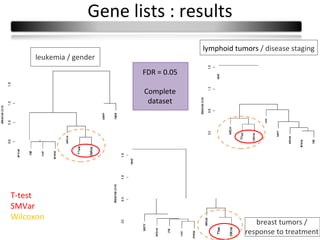
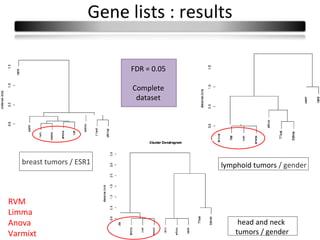
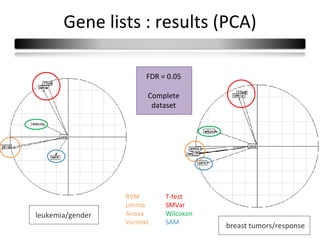
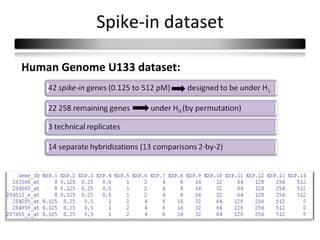
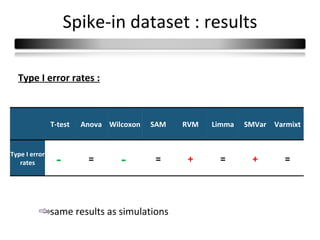
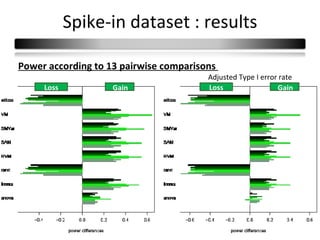
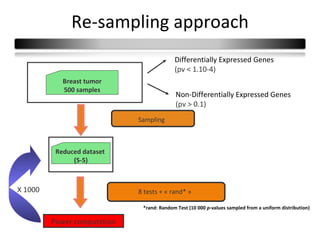
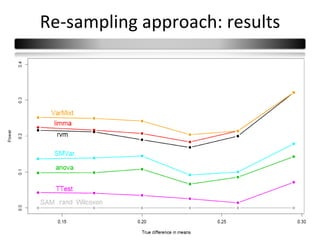
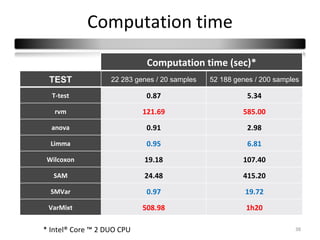
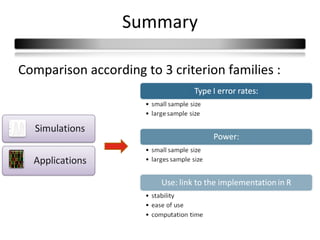
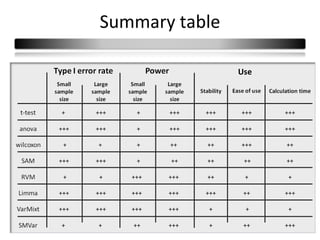
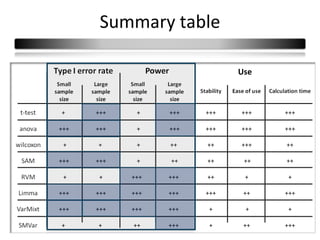
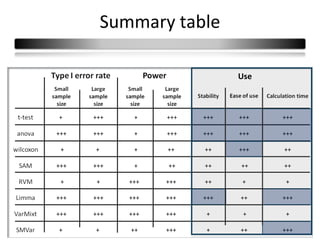
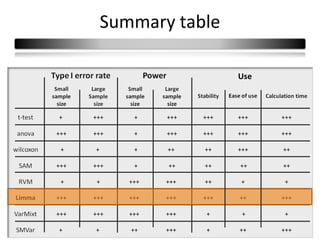
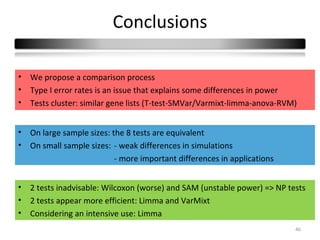
This document summarizes a study that compares 8 different statistical tests for differential gene expression analysis on microarray data. The study uses simulations with different data models to evaluate the power and performance of each test under various conditions. The tests are applied to the simulated data to identify differentially expressed genes and their ability to correctly detect differentially expressed genes is assessed and compared across the different simulation models.

![Variance modelling
An essential point: variance modelling
«…accurate estimation of variability is difficult. » (RVM, Wright and Simon, 2003)
« The importance of variance modelling is now widely known…» (SMVar, Jaffrézic et al, 2007)
« Many different sources of variability affect gene expression intensity measurements […]. Not at
all are well characterized or even identified» (VarMixt, Delmar et al, 2004)
Many approaches :
6](https://image.slidesharecdn.com/b4-jeanmougin-120405061500-phpapp02/85/B4-jeanmougin-6-320.jpg)