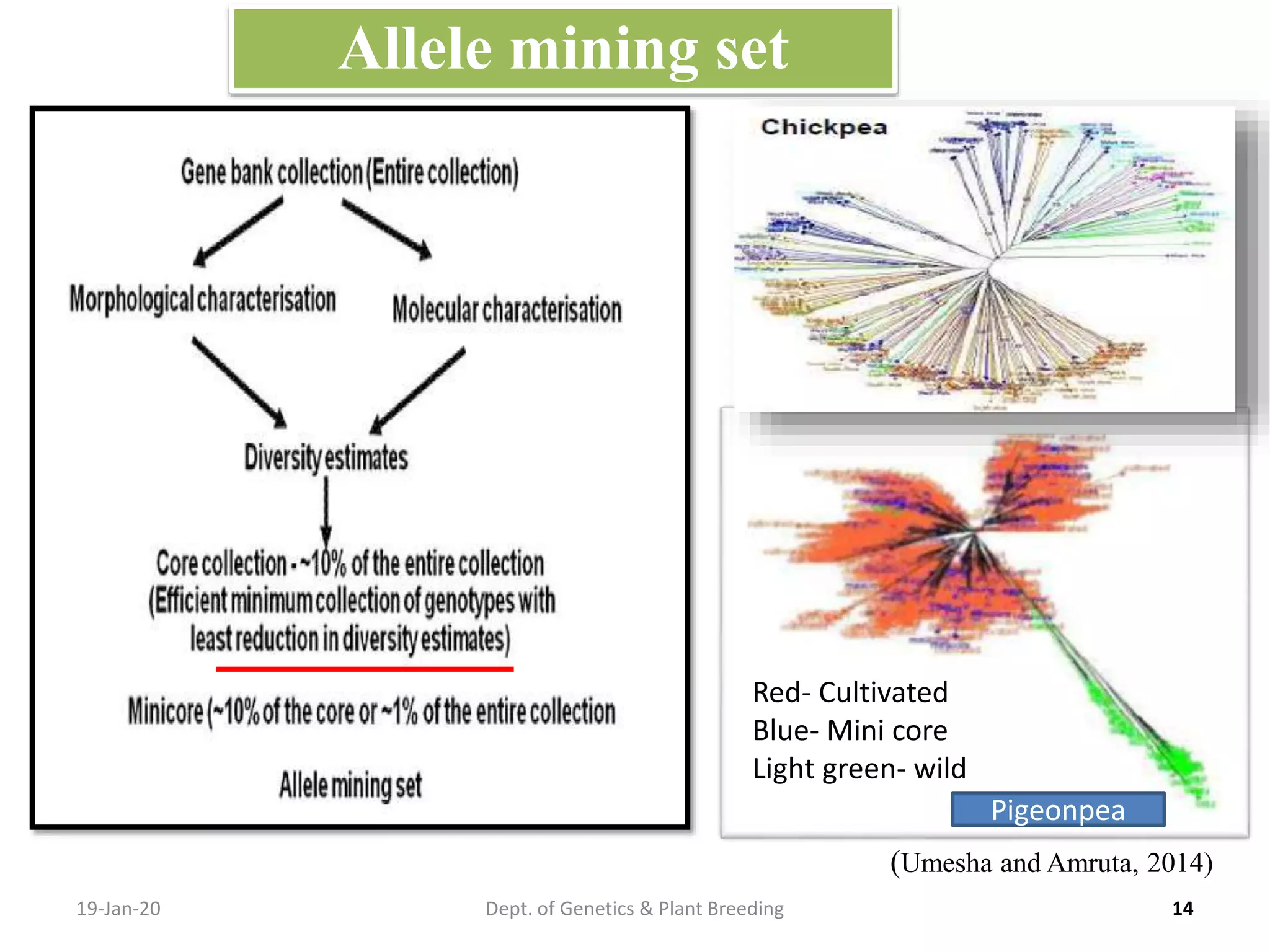
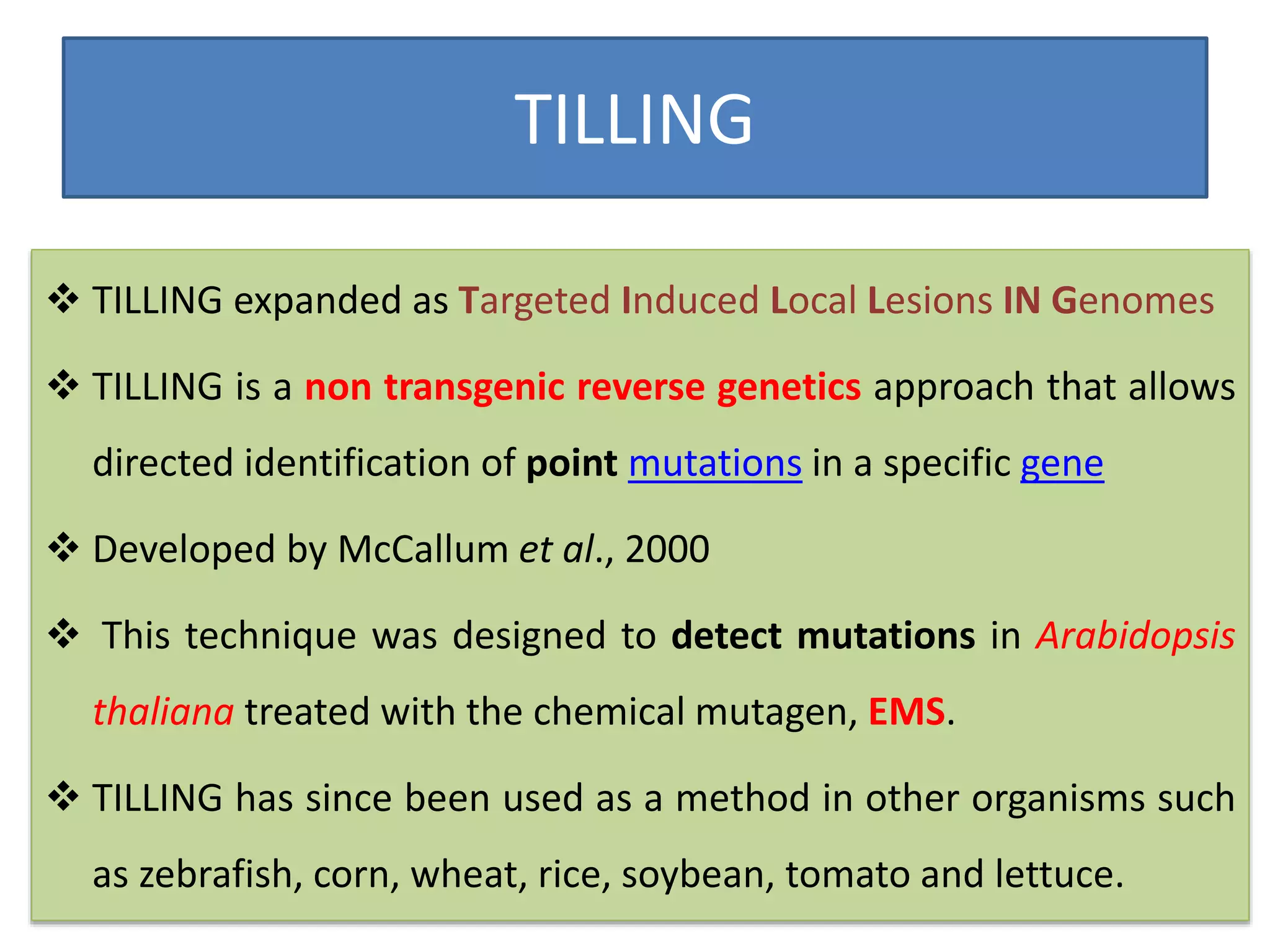
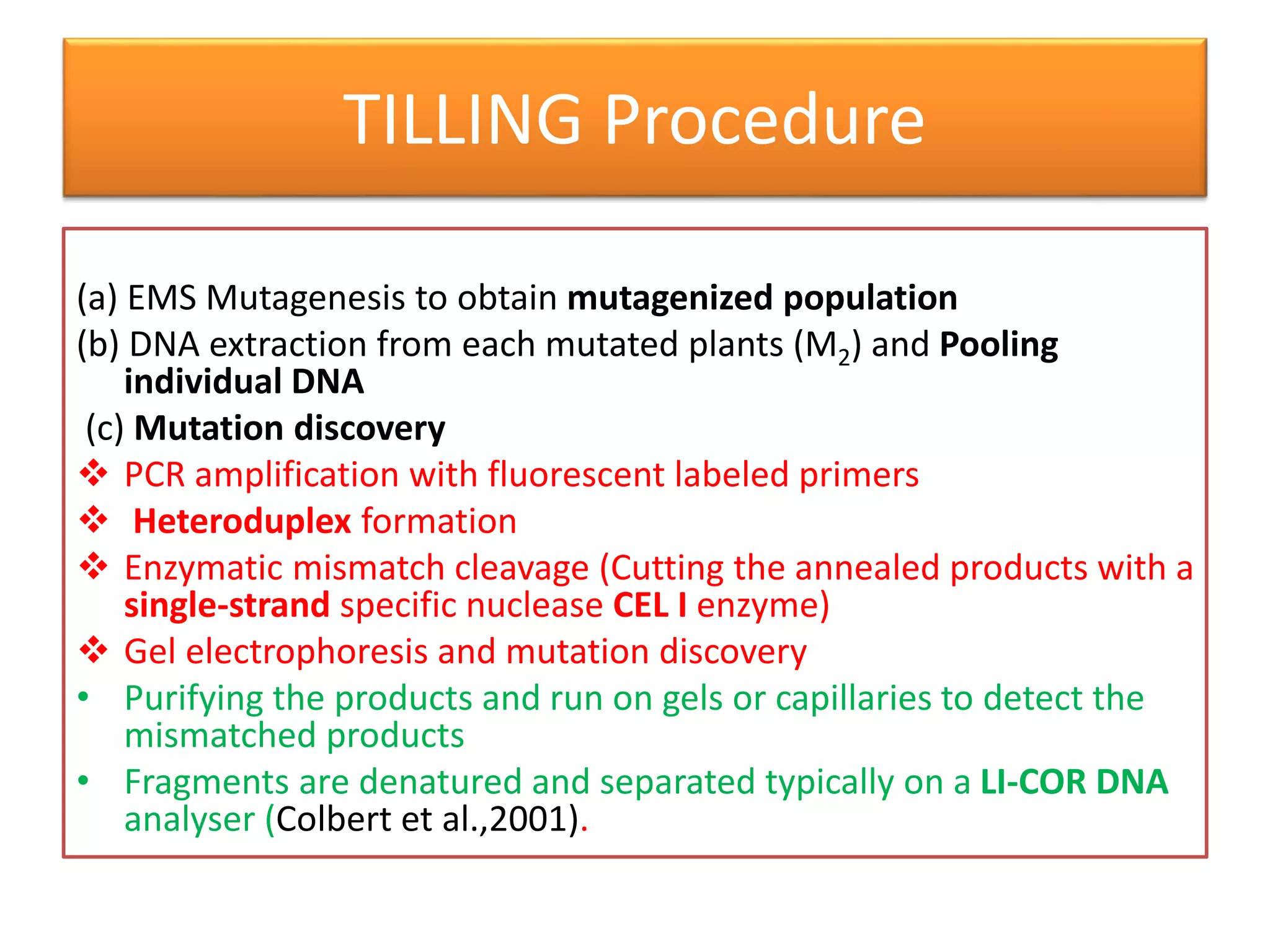
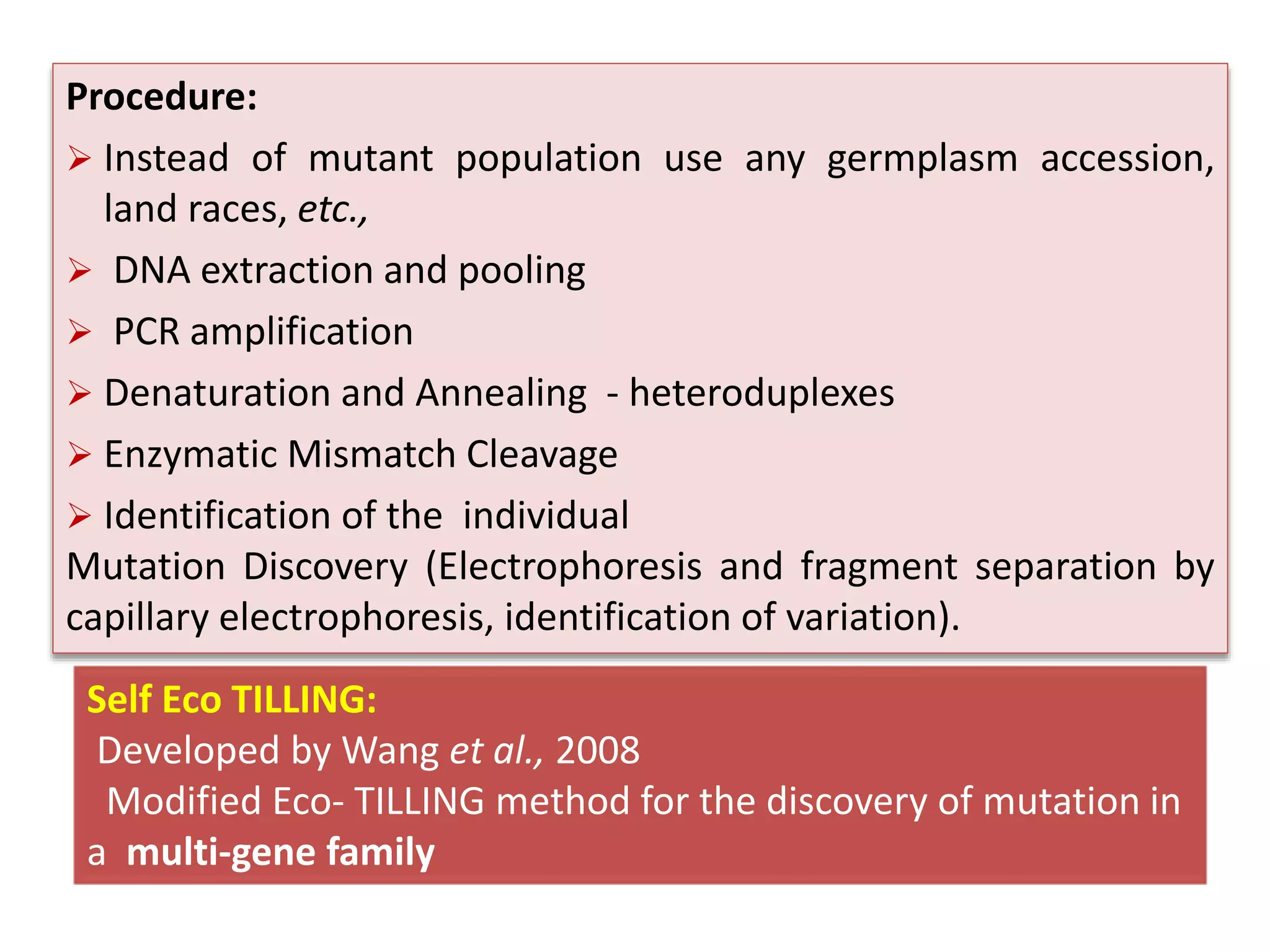
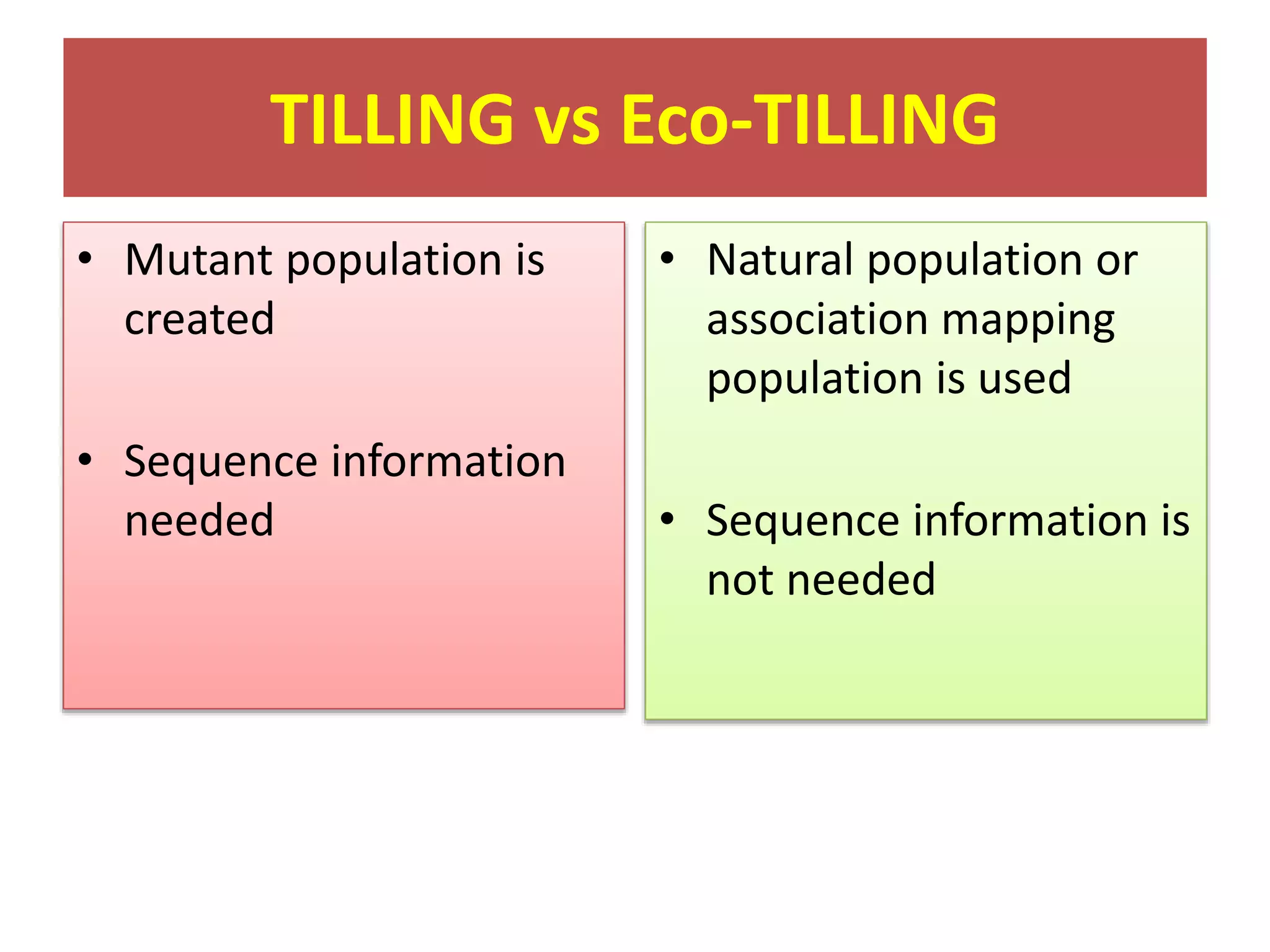
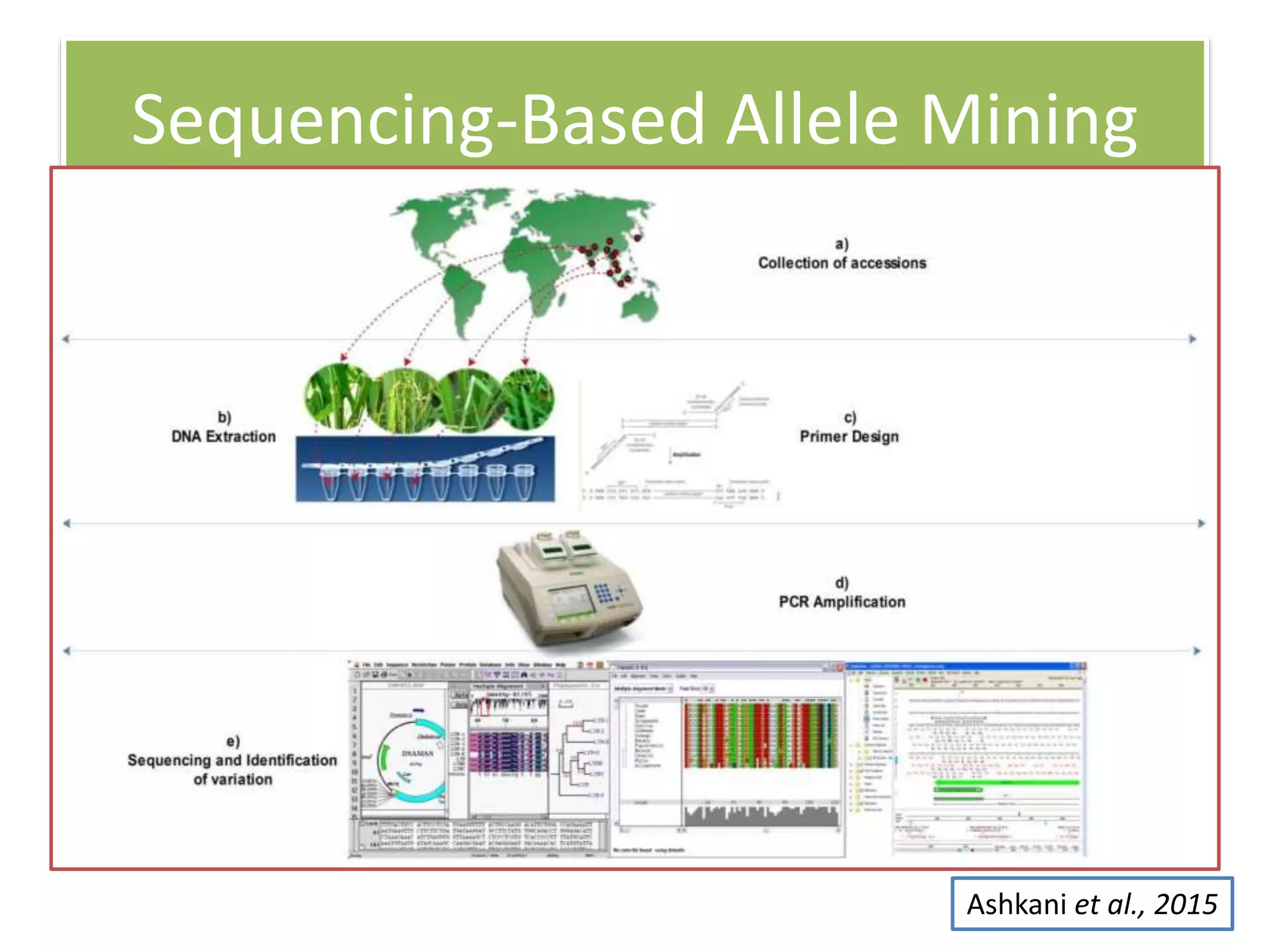
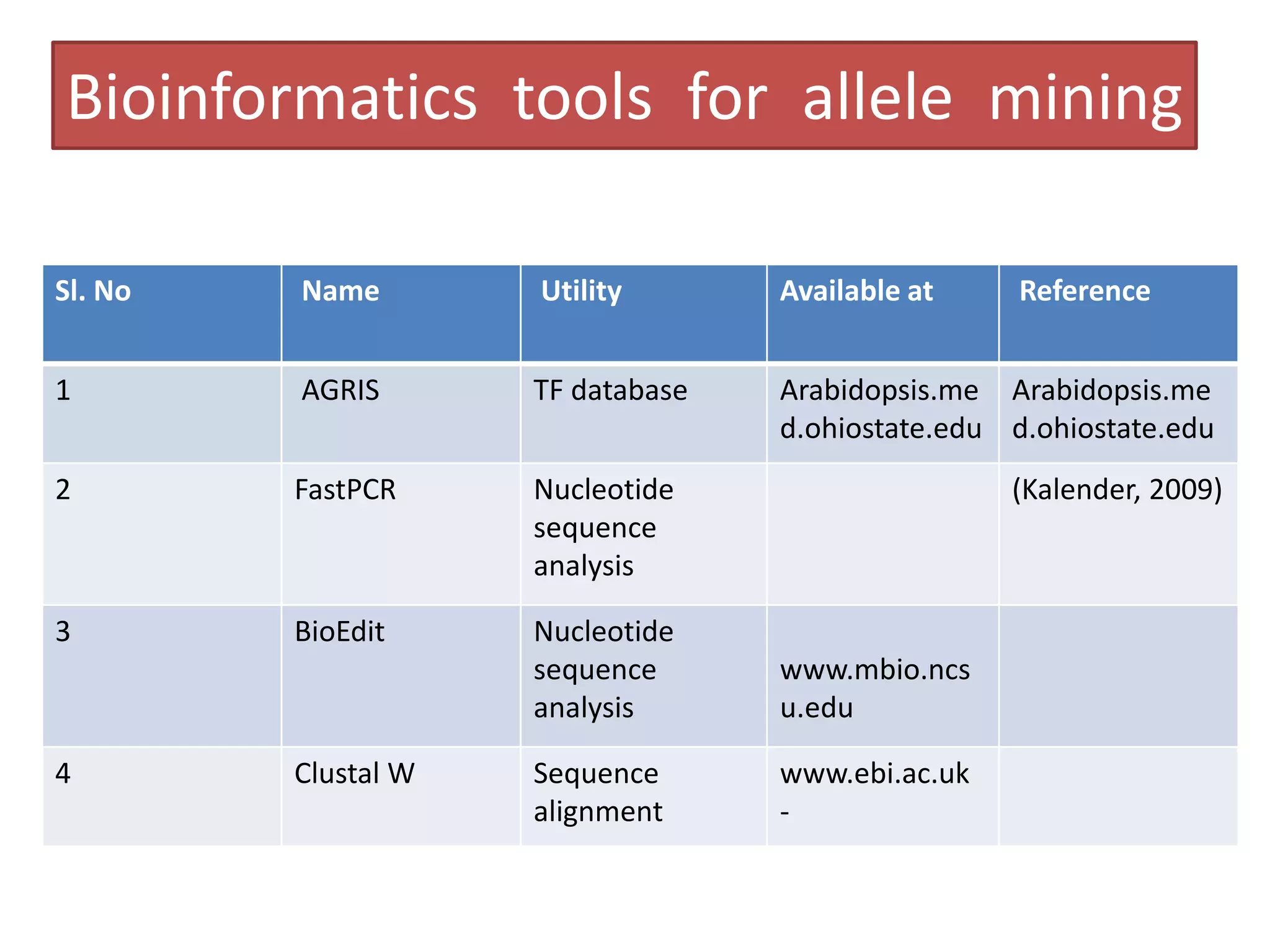
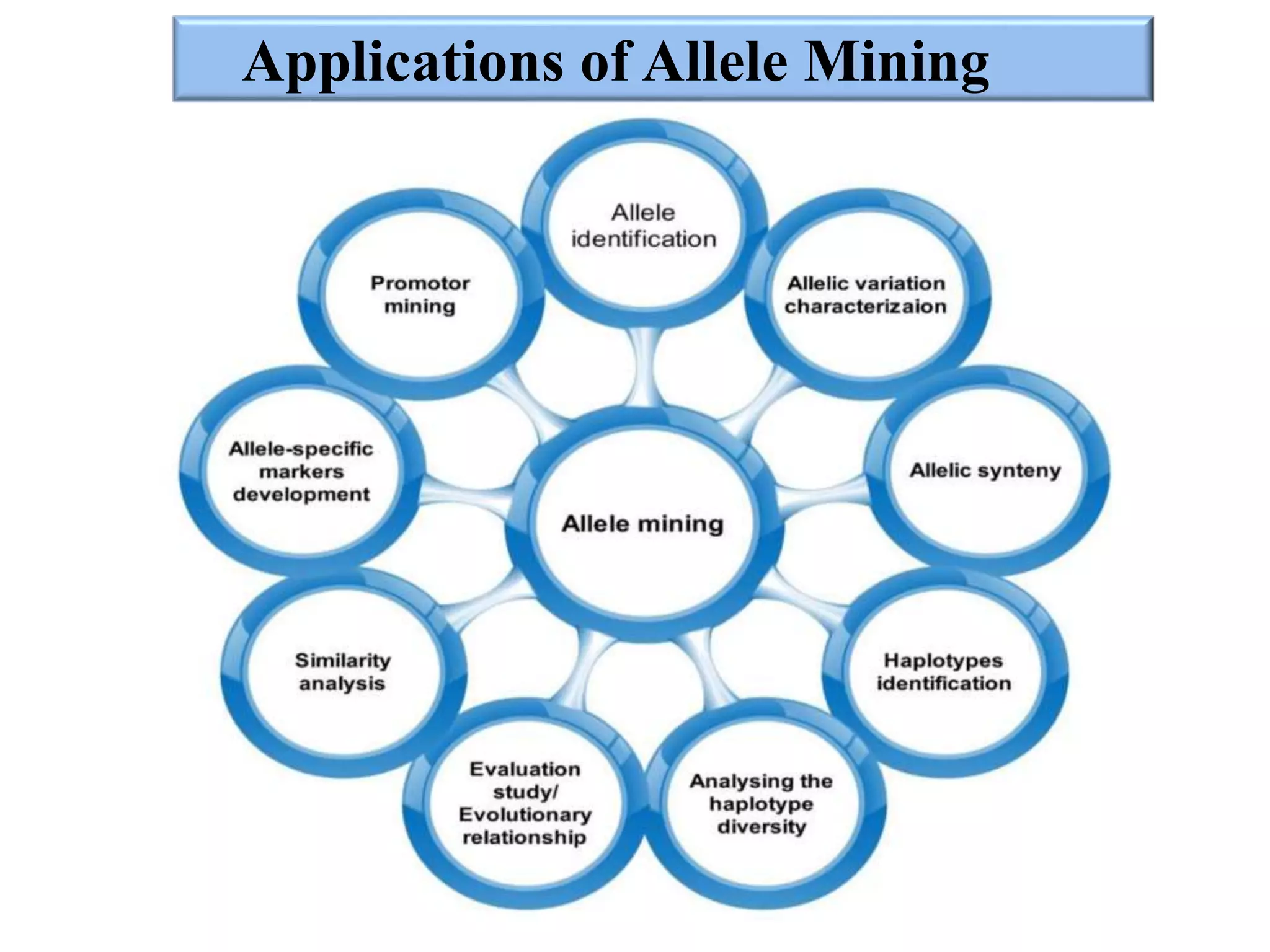
This document discusses allele mining as a technique for improving crops. It defines allele mining as identifying allelic variation within genetic resources collections to find superior alleles. There are two main approaches - TILLING based allele mining which uses mutagenized populations and enzymatic cleavage to find mutations, and sequencing-based allele mining which uses PCR and sequencing to identify natural variation. Both have benefits and limitations. Applications of allele mining include finding alleles for resistance, abiotic stress tolerance, and improved yield and quality. Overall, allele mining is a promising approach for utilizing genetic resources to discover variants that can aid crop breeding.