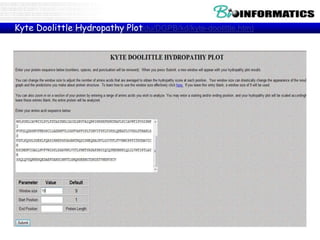
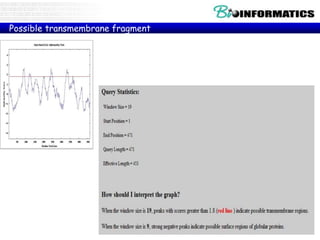
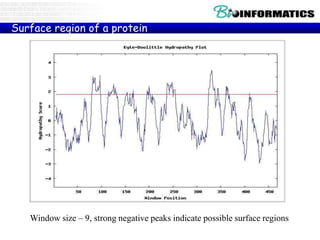
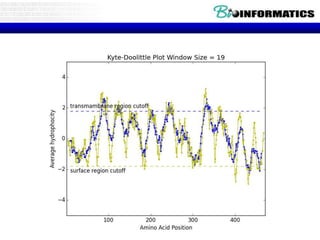
The document discusses hosting a GIT repository on GitHub and accepting an invitation to a shared repository called "Bioinformatics-I-2015" hosted on GitHub.ugent.be. It also lists some example control structures for Python scripts like if/else statements and for loops and poses extra bioinformatics analysis questions that could be answered using Python scripts.



![Control Structures
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue](https://image.slidesharecdn.com/2015bioinformaticsbiopythonpart4-151130225843-lva1-app6891/85/2015-bioinformatics-bio_python_part4-4-320.jpg)

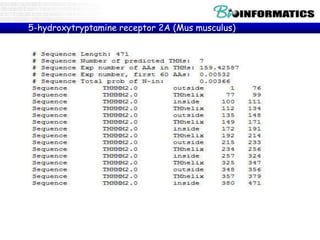
![5-hydroxytryptamine receptor 2A isoform 1 [Homo sapiens]
NCBI Reference Sequence: NP_000612.1
GenPept Identical Proteins Graphics
>gi|10835175|ref|NP_000612.1| 5-hydroxytryptamine receptor 2A isoform 1 [Homo sapiens]
MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLH
LQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYR
WPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPI
PVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLAS
FSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICK
ESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYK
SSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV](https://image.slidesharecdn.com/2015bioinformaticsbiopythonpart4-151130225843-lva1-app6891/85/2015-bioinformatics-bio_python_part4-8-320.jpg)