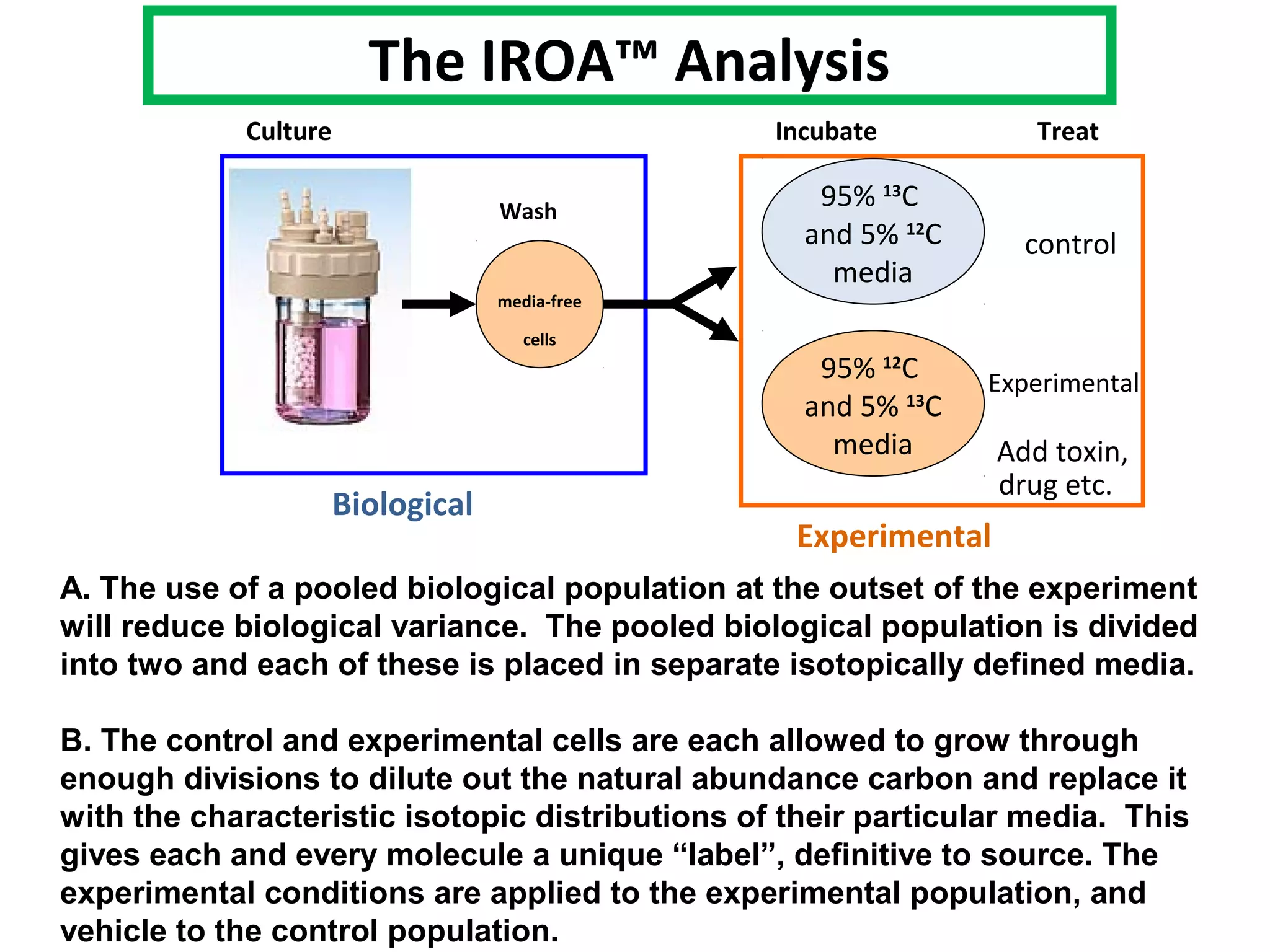
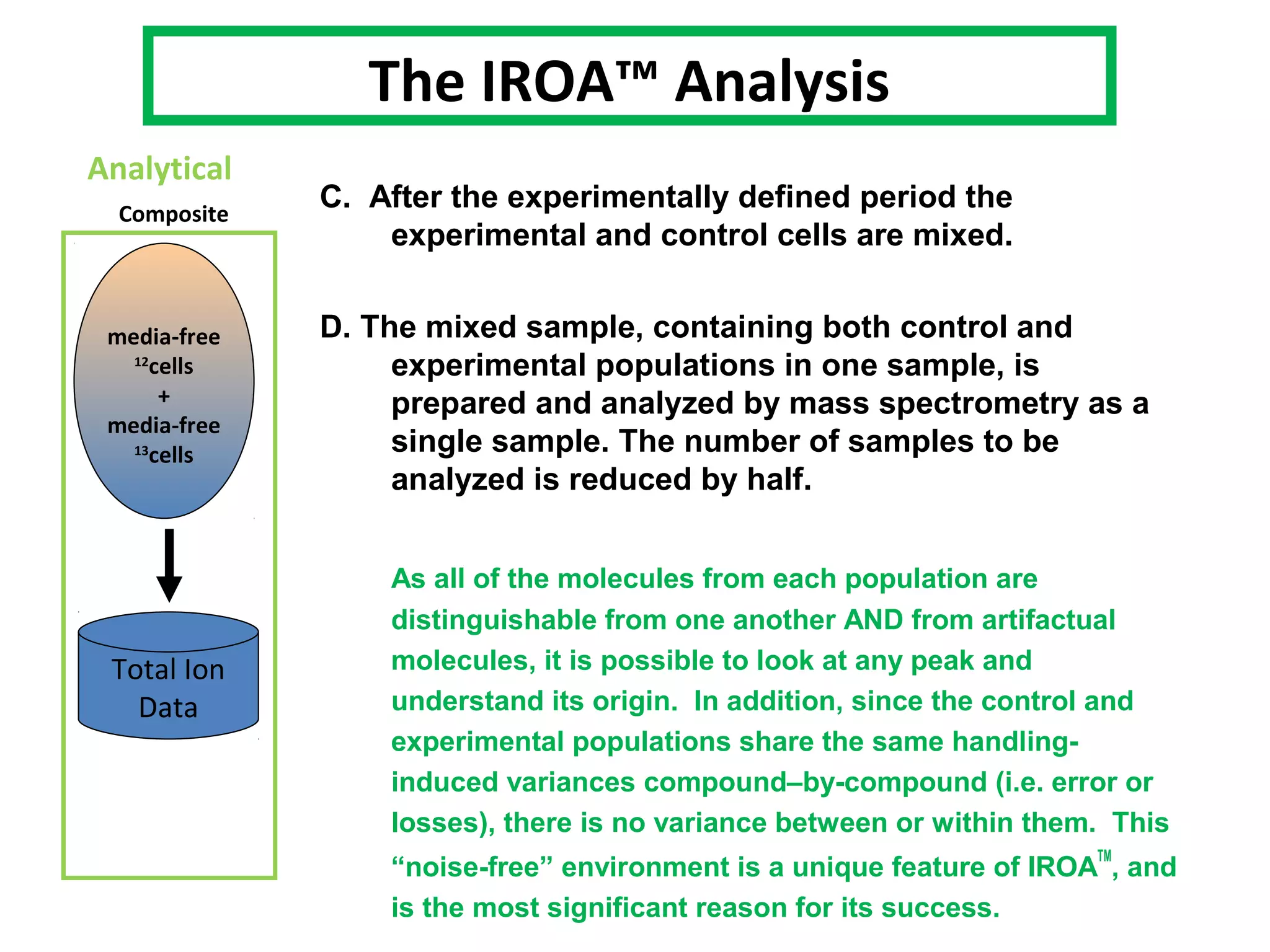
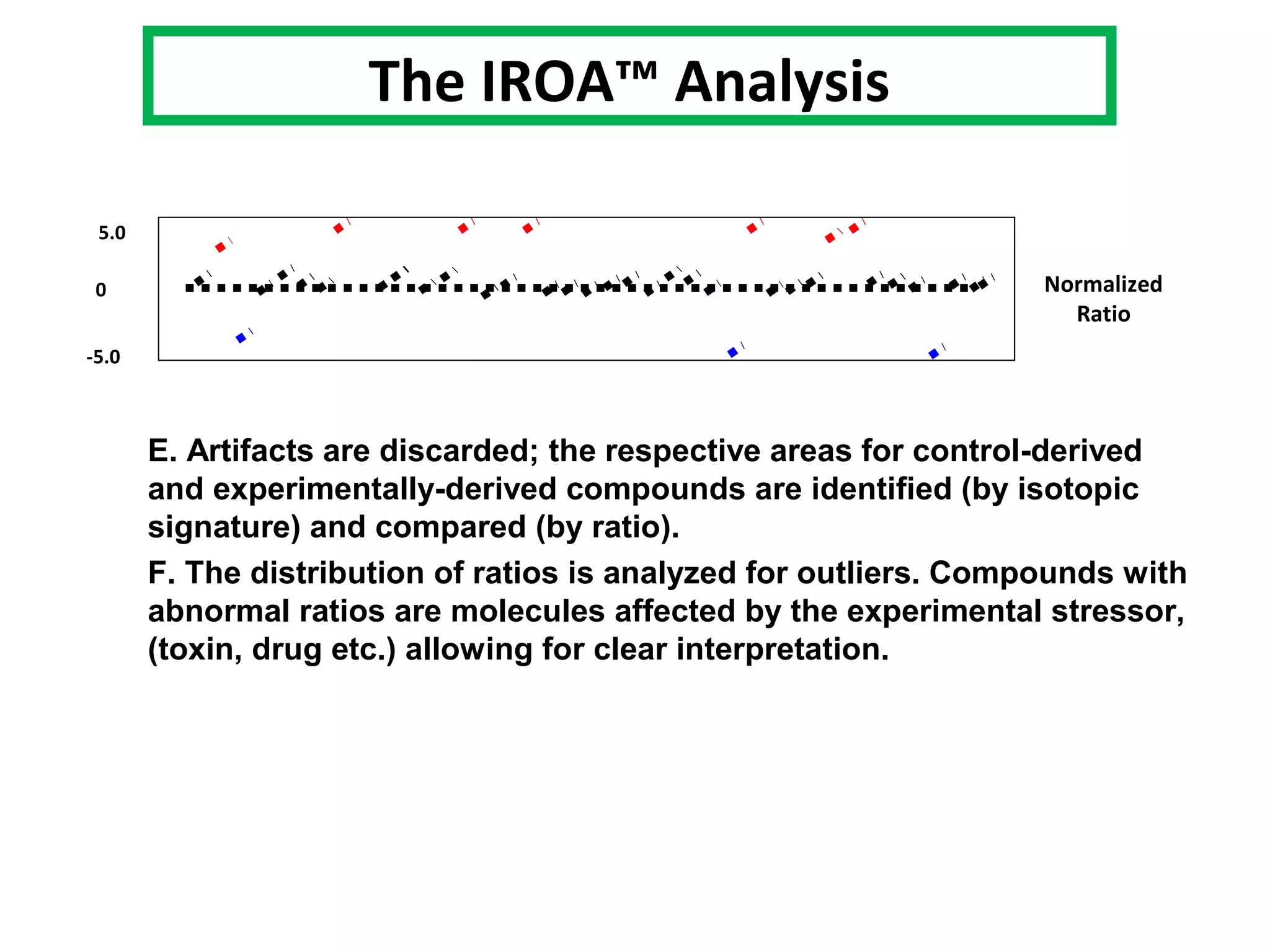
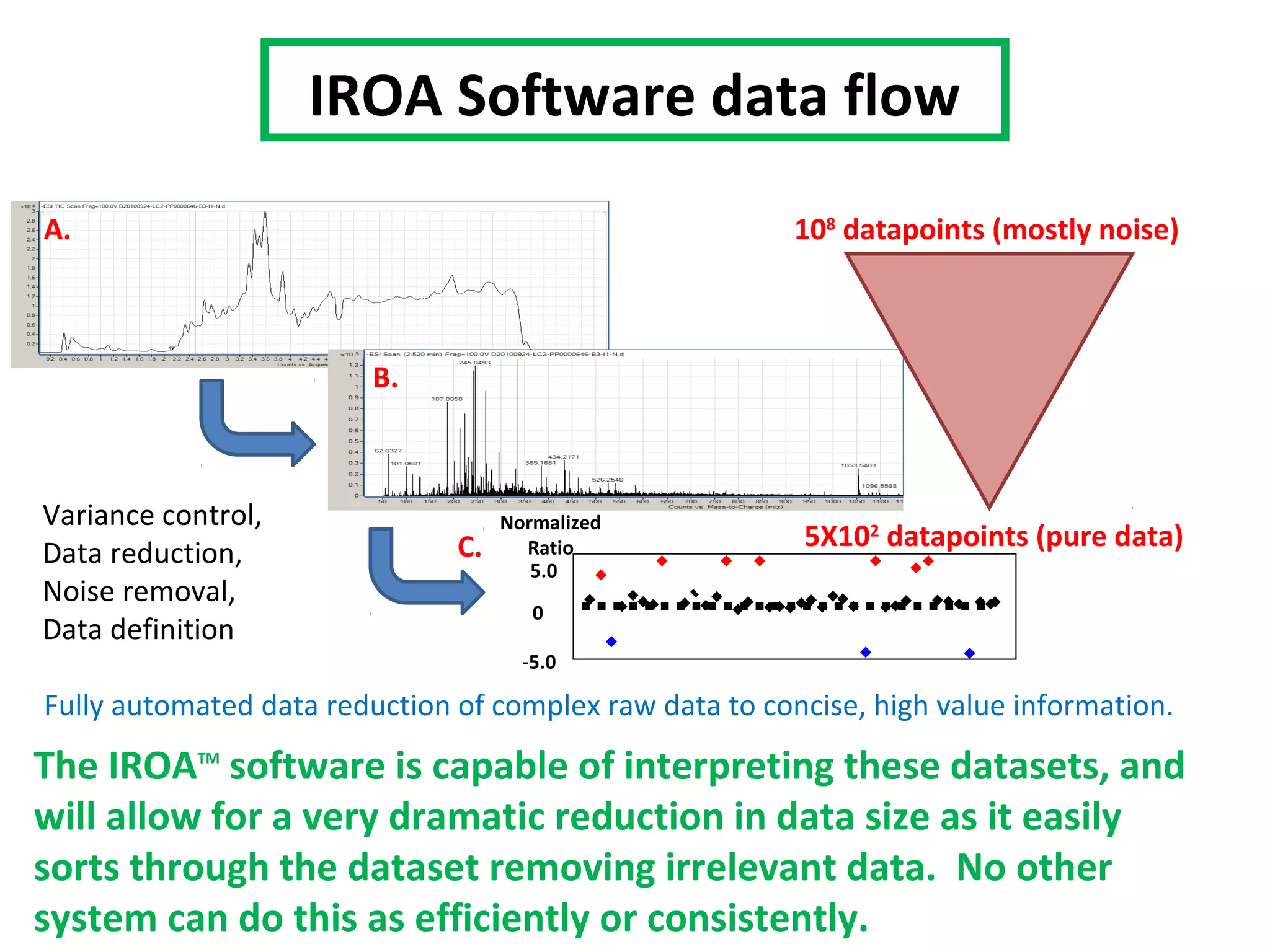
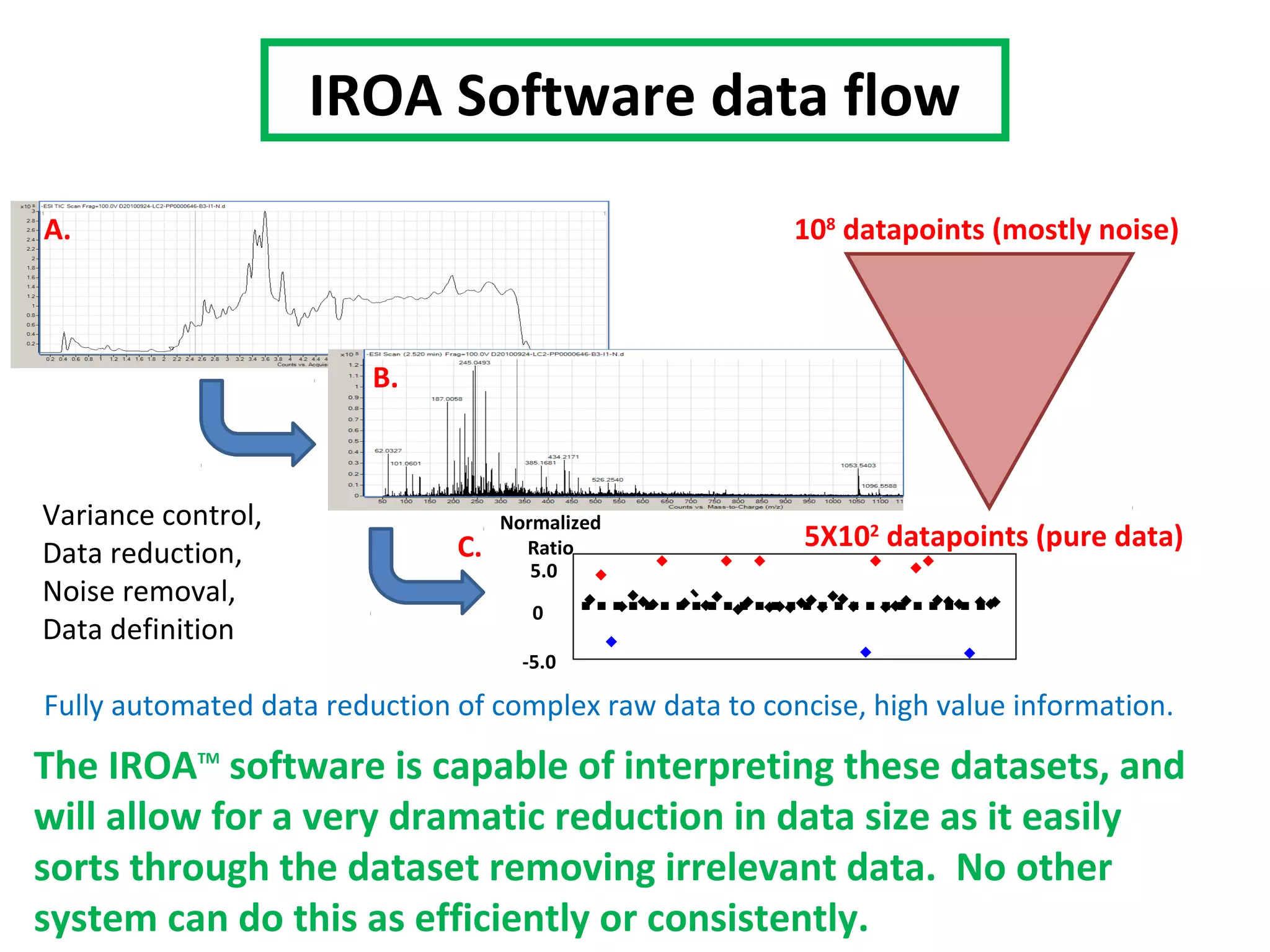
The IROATM methodology uses isotopically labeled media to automatically simplify metabolomics analysis. It requires isotopically paired media and software to extract data from mass spectrometry files. The method involves growing control and experimental cell populations in different isotopic media to label molecules. The populations are then mixed and analyzed together as one sample by mass spectrometry. Software then identifies control and experimental molecules based on isotopic signatures and detects outliers affected by experimental stressors.