gEVAL - A Genome Evaluation Browser for Improving Genome Assemblies (SFAF 2014 Poster)
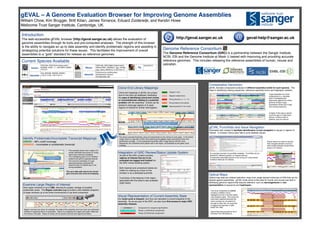
- 1. gEVAL – A Genome Evaluation Browser for Improving Genome Assemblies! William Chow, Kim Brugger, Britt Kilian, James Torrance, Eduard Zuiderwijk, and Kerstin Howe! Wellcome Trust Sanger Institute, Cambridge, UK.! http://geval.sanger.ac.uk! geval-help@sanger.ac.uk! gEVAL Punchlists and Issue Navigation! Automated lists created to facilitate identification of and navigation to issues or regions of interest. In browser menus also help to jump between issues.! ! Optical Maps! Optical map data are ordered restriction maps from single stained molecules of DNA that can be aligned against assemblies. gEVAL hosts some of this data for human and mouse and aids in identifying genomic regions that requires attention, such as rearrangements or mis-representation of sequence and haplotypes. ! ! Introduction! The web-accessible gEVAL browser (http://geval.sanger.ac.uk) allows the evaluation of genome assemblies through its tools and pre-computed analyses. The strength of this browser is the ability to navigate an up to date assembly and identify problematic regions and assisting in strategizing potential solutions for these issues. This facilitates the improvement of overall assemblies to a “gold” standard for release as reference genomes.! Mapped multiple times Wrong direction (<<, <>, >>) Wrong distance from partner Integration of GRC Review/Status Update System! A! s part of the GRC curation process, regions of interest that are to be evaluated are tagged and tracked via the GRC review ticketing system. ! ! Both resolved and unresolved tickets are visible for viewing as a track on the browser or as a dedicated punchlist. ! ! A summary of the features in the region associated with the ticket is also available (right insert).! Visual Representation of Current Assembly State! Our build cycle is frequent, and thus can represent a current snapshot of the assembly. As we are part of the GRC, we also have first access to major GRC assembly releases.! Component in sequencing Pipeline. Phase 1 unfinished component. Phase 2/3 finished component. Comparative Genomics! gEVAL includes comparative analyses of different assembly builds for each species. This helps in identifying missing sequences, reference assembly errors and haplotypic variation.! ! A gap separates two clone components in a zebrafish bulid. Investigating the alignments against two whole genome shotgun (wgs) assemblies reveal size of gap and missing sequence.! A region of the wgs is used to cover the gap in a later build. (bonus: a clone is also in pipeline, grey box above). ! The clone component AL596089 contains a deletion and is highlighted by the 3 cell line optical map analysis (right). This would not have been captured because the clone overlaps do not extend far enough to show this. An issue that is tagged and reported in GRC ticket: HG-1482.! ! Optical Map data provided by the D. Schwartz Lab (UW Madison).! Popup menus on tracks to quickly help navigate between previous/ next overlap between components along a chr (below).! An example overview of punchlists available. Punchlists can be tailored for different projects, on request (above).! Components potentially placed on the wrong chr using marker evidence listed per chr (below).! Current Species Available! Identify Problematic/Incomplete Transcript Mappings! GREEN – 98% cutoff coverage ! ORANGE – Incomplete or problematic transcript! ! • This example shows how a region of 2 clones (dark/light blue boxes on contig track) have incorrect orientation.! • The overlapping gene ryr1b, therefore looked to be split on opposite strands! • The incorrect orientation of 2 gap spanning fosmids confirmed the assertion that CU138549 was in the wrong orientation.! ! The up to date path returns the correct gene structure and clone end mapping.! before! after! Examine Large Region of Interest! View region windows of up to 2Mb, allowing for greater vantage of possible problematic areas. The Region overview page provides a less detailed snapshot of larger windows up to the entire chromosome or top level component.! ! ! Region overview can show, for example, the state of the assembly and how much are unfinished, finished or sequence that is in production. Above is a snapshot of a region just under 10Mb and the clones in the path. Status of clones can be quickly scanned and regions prioritized. ! Clone End Library Mappings! ! Mapped 1 time Spanning partner in the vicinity Clone end mappings in gEVAL are unique due to how they are displayed, facilitating the ease of identifying concurrent clones or inconsistencies relating to a potential problem with the assembly. Clones can be picked to close gap regions or to span regions of interest for further interrogation.! ! before! after! The above example illustrates using end placements to pick clones to cover gaps. In the before image, there is a gap with a BAC clone spanning the gapped region according to their end placements (orange). In the subsequent assembly (after image above) with the clone sequenced, the unfinished clone places well in the region, as illustrated by the green clone overlaps.! Human! GRCh38, GRCh37pX (latest patch), NCBI36, CHM1_1.1, NA12878, HuREF, YH1/2.0. ! Zebrafish! Zv9, WGS28, WGS29, WGS31, ! z.2013.12.06, z.2014.03.14.! ! Mouse! GRCm38, GRCm38pX (latest patch), GRCm37B/C, NCBIm37, wgs_c57bl6j, wgs_celera, MGSCv3, m.2013.03.15.! Helminth! Echinococcus multilocularis! Schistosoma mansoni ! Stronglyoides ratti! Genome Reference Consortium! The Genome Reference Consortium (GRC) is a partnership between the Sanger Institute, NCBI, EBI and the Genome Institute at Wash U tasked with improving and providing accurate reference genomes. This includes releasing the reference assemblies of human, mouse and zebrafish. ! Pig! Sscrofa10.2! The red arrows highlights the incorrect orientation of these ryr1b gene split fosmid ends. ! on opposite strands! Clone end placements reveal sequence that can be placed in the gap region. Assembly reveals newly sequenced clone in path.!
