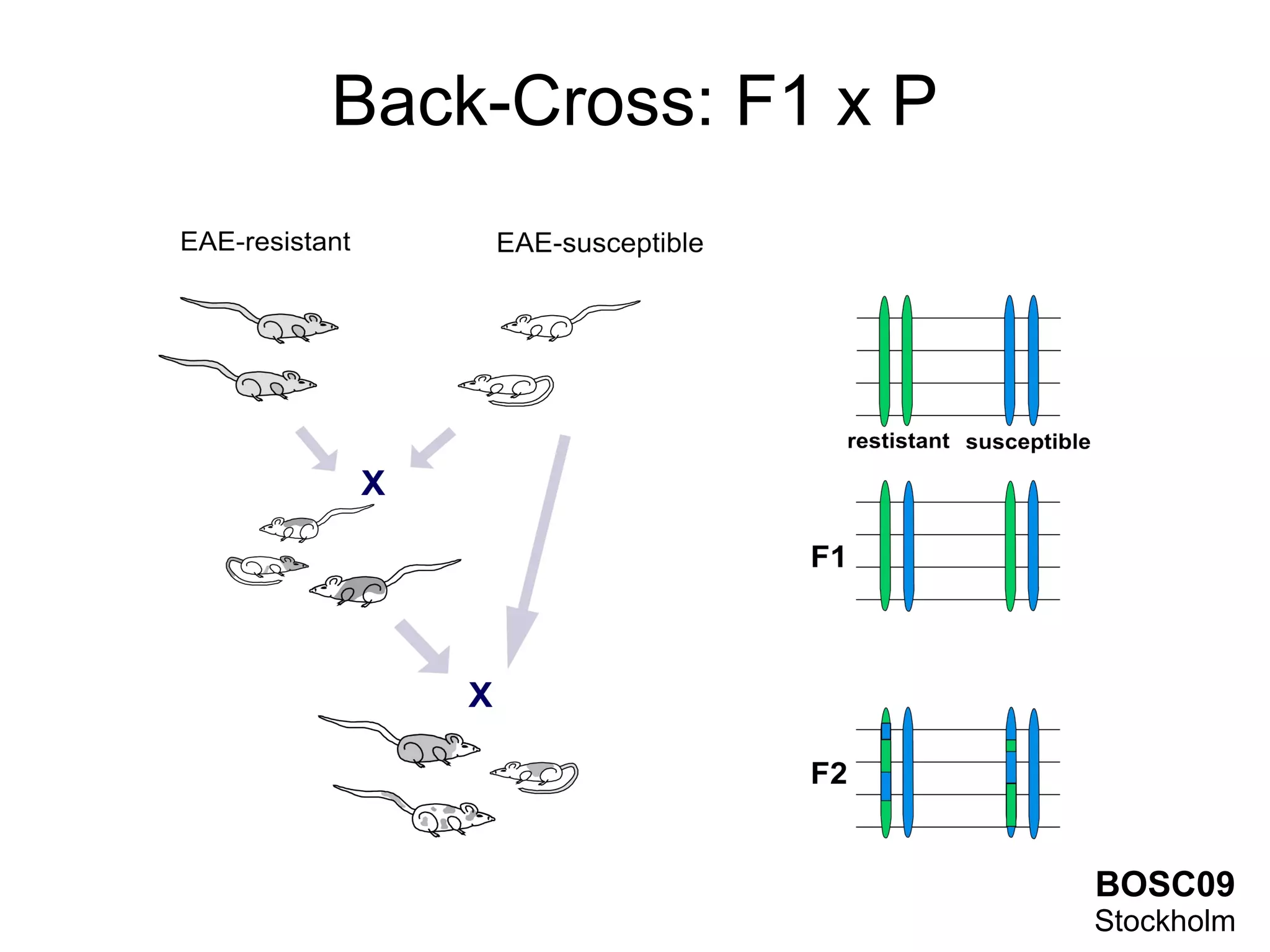
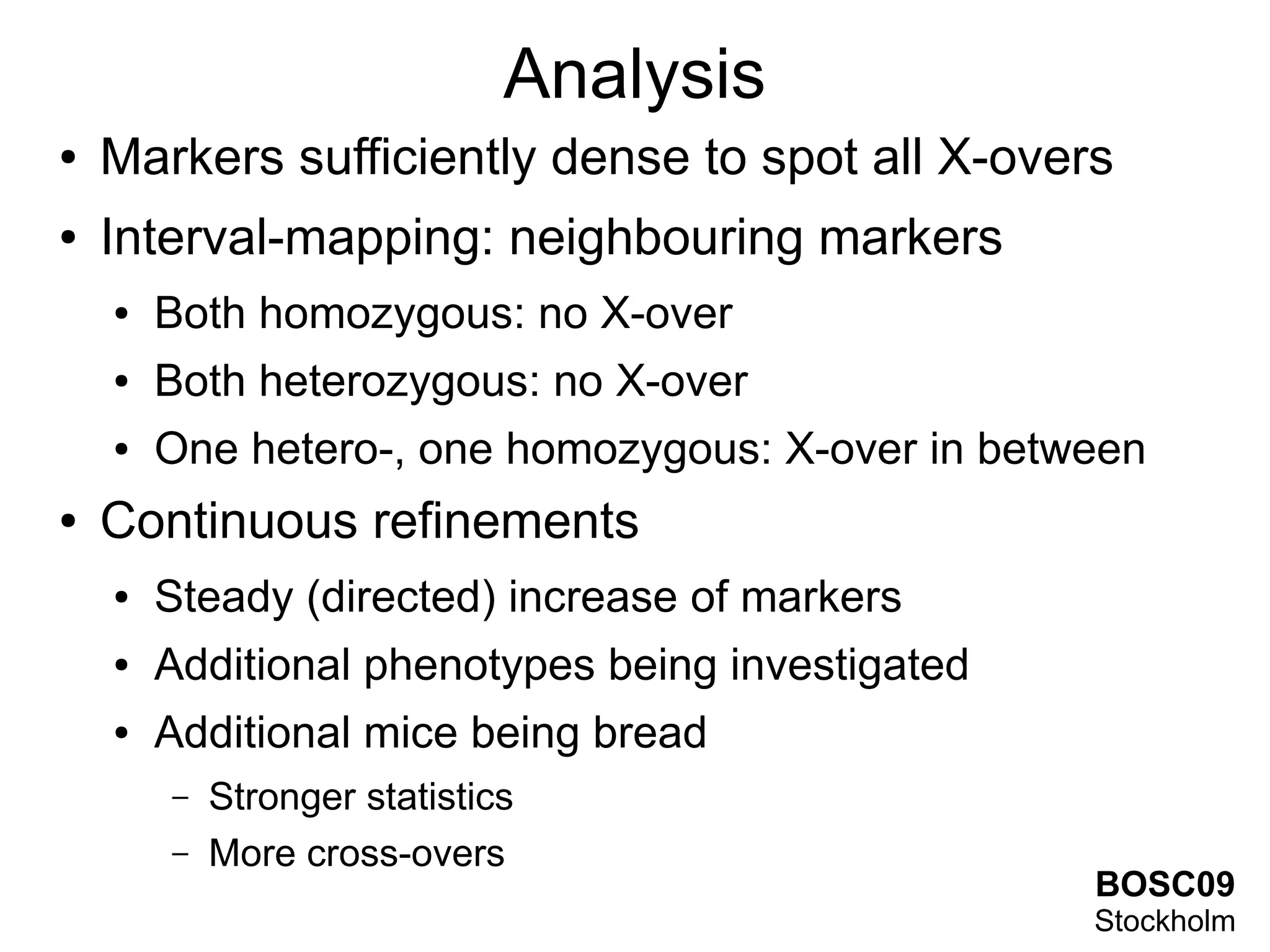
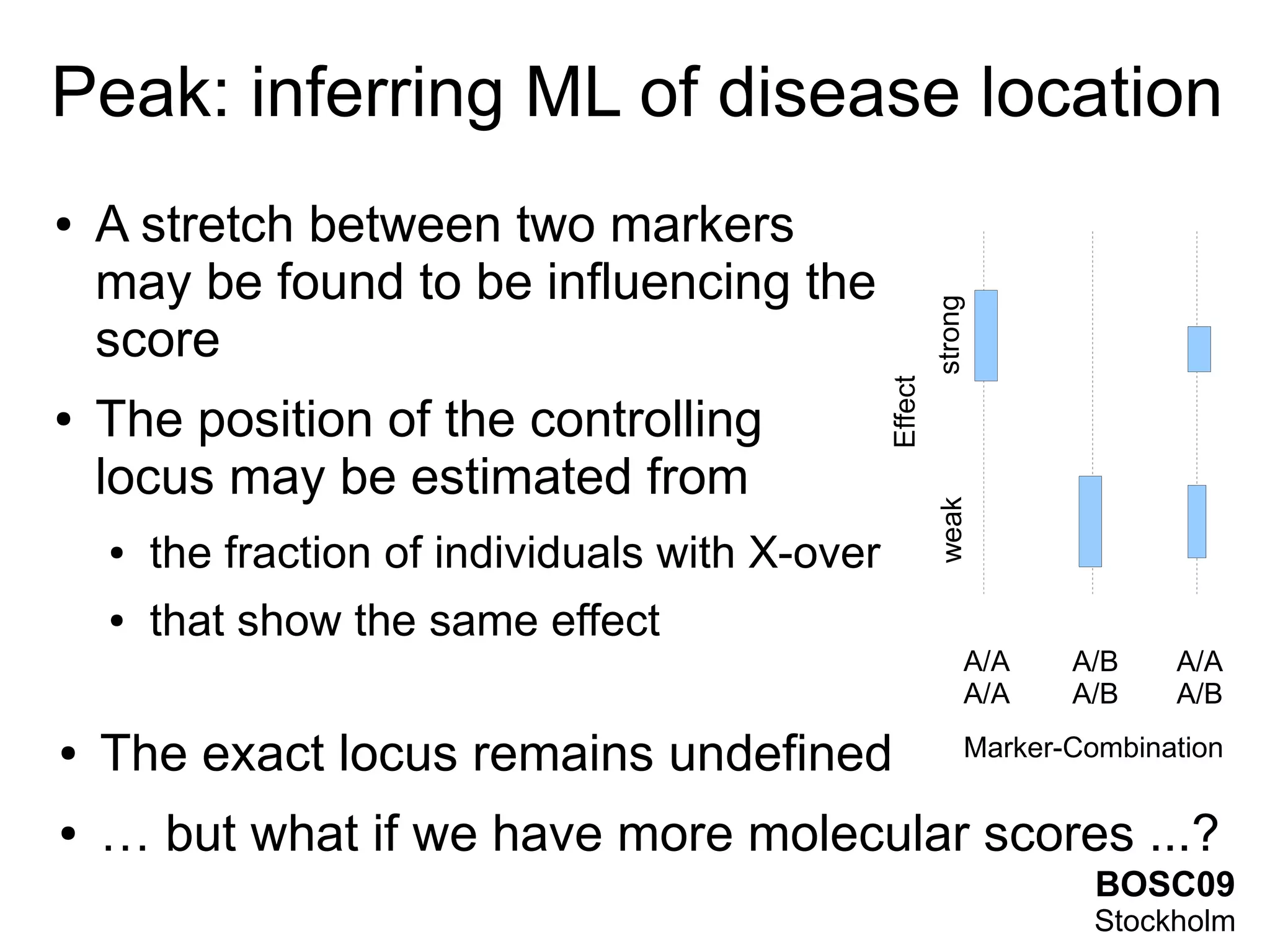
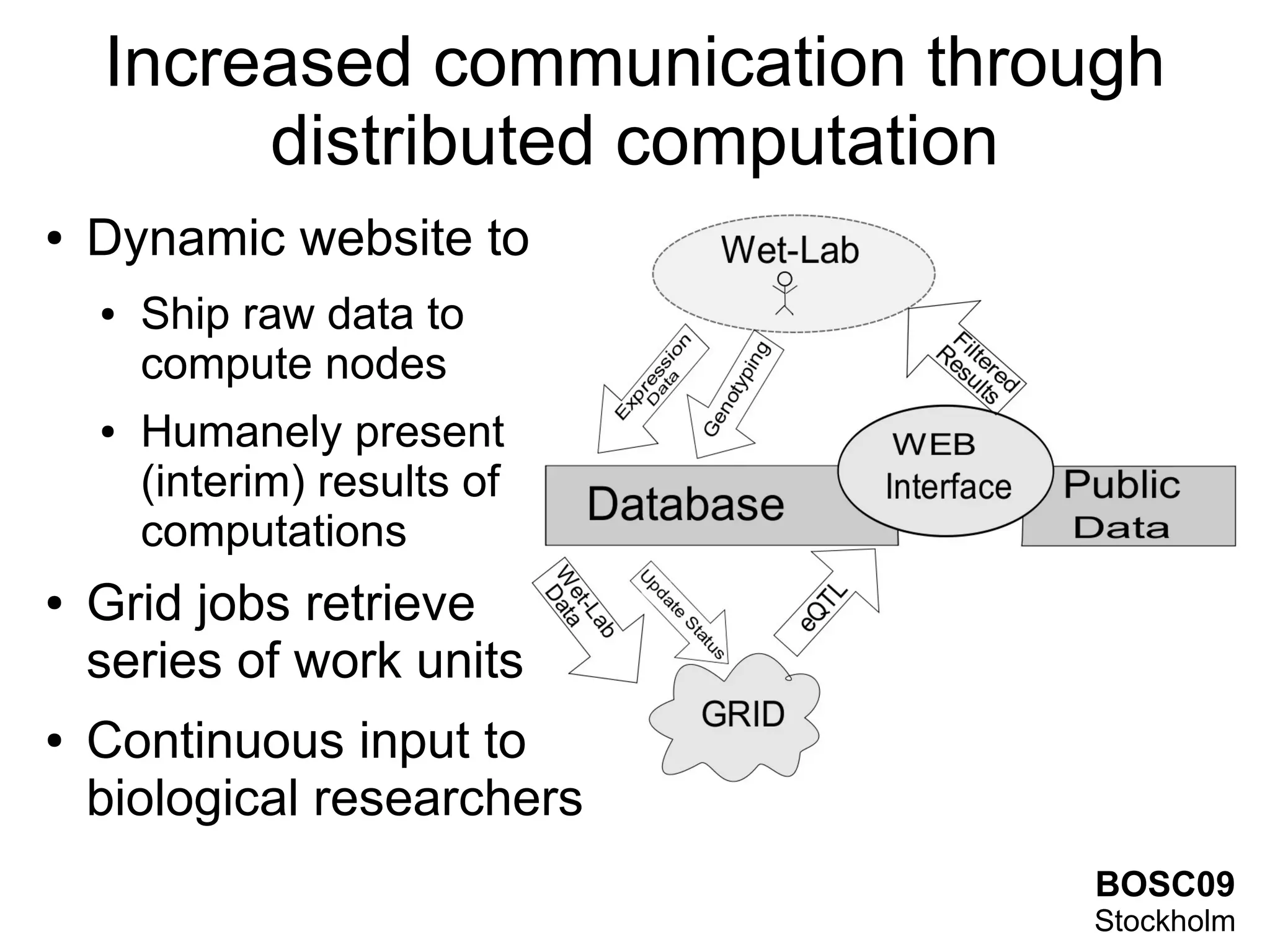
This document summarizes grid-based expression QTL analysis. It discusses isolating susceptible or resistant strains of an organism to a disease, identifying genetic markers that differ between strains, generating offspring with mixed genotypes to score disease phenotypes, and using statistical methods to determine associations between markers and scores. It describes using dense genetic markers and interval mapping to localize where crossovers occur on chromosomes. The analysis can use gene expression levels as scores and link genetic loci to controlled genes and pathways. Distributed computation is proposed to handle the large amount of data generated from thousands of genes, genetic markers, individuals, and phenotypes.